A Multicomponent Neuronal Response Encodes the Larval Decision to Pupariate upon Amino Acid Starvation
- PMID: 30301757
- PMCID: PMC6246885
- DOI: 10.1523/JNEUROSCI.1163-18.2018
A Multicomponent Neuronal Response Encodes the Larval Decision to Pupariate upon Amino Acid Starvation
Erratum in
-
Erratum: Jayakumar et al., "A Multicomponent Neuronal Response Encodes the Larval Decision to Pupariate upon Amino Acid Starvation".J Neurosci. 2020 Mar 4;40(10):2178-2181. doi: 10.1523/JNEUROSCI.2713-19.2019. Epub 2020 Feb 27. J Neurosci. 2020. PMID: 32107276 Free PMC article. No abstract available.
Abstract
Organisms need to coordinate growth with development, particularly in the context of nutrient availability. Thus, multiple ways have evolved to survive extrinsic nutrient deprivation during development. In Drosophila, growth occurs during larval development. Larvae are thus critically dependent on nutritional inputs; but after critical weight, they pupariate even when starved. How nutrient availability is coupled to the internal metabolic state for the decision to pupariate needs better understanding. We had earlier identified glutamatergic interneurons in the ventral ganglion that regulate pupariation on a protein-deficient diet. Here we report that Drosophila third instar larvae (either sex) sense arginine to evaluate their nutrient environment using an amino acid transporter Slimfast. The glutamatergic interneurons integrate external protein availability with internal metabolic state through neuropeptide signals. IP3-mediated calcium release and store-operated calcium entry are essential in these glutamatergic neurons for such integration and alter neuronal function by reducing the expression of multiple ion channels.SIGNIFICANCE STATEMENT Coordinating growth with development, in the context of nutrient availability is a challenge for all organisms in nature. After attainment of "critical weight," insect larvae can pupariate, even in the absence of nutrition. Mechanism(s) that stimulate appropriate cellular responses and allow normal development on a nutritionally deficient diet remain to be understood. Here, we demonstrate that nutritional deprivation, in postcritical weight larvae, is sensed by special sensory neurons through an amino acid transporter that detects loss of environmental arginine. This information is integrated by glutamatergic interneurons with the internal metabolic state through neuropeptide signals. These glutamatergic interneurons require calcium-signaling-regulated expression of a host of neuronal channels to generate complex calcium signals essential for pupariation on a protein-deficient diet.
Keywords: Drosophila; SOCE; glutamatergic neurons; nutrient-sensing.
Copyright © 2018 Jayakumar et al.
Figures
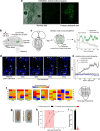



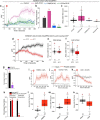

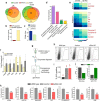
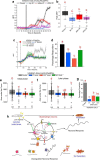
Similar articles
-
IP3-mediated Ca2+ signals regulate larval to pupal transition under nutrient stress through the H3K36 methyltransferase Set2.Development. 2021 Jun 1;148(11):dev199018. doi: 10.1242/dev.199018. Epub 2021 Jun 12. Development. 2021. PMID: 34117888
-
Drosophila larval to pupal switch under nutrient stress requires IP3R/Ca(2+) signalling in glutamatergic interneurons.Elife. 2016 Aug 5;5:e17495. doi: 10.7554/eLife.17495. Elife. 2016. PMID: 27494275 Free PMC article.
-
Store-Operated Calcium Entry through Orai Is Required for Transcriptional Maturation of the Flight Circuit in Drosophila.J Neurosci. 2015 Oct 7;35(40):13784-99. doi: 10.1523/JNEUROSCI.1680-15.2015. J Neurosci. 2015. PMID: 26446229 Free PMC article.
-
Surviving nutritional deprivation during development: neuronal intracellular calcium signaling is critical.Int J Dev Biol. 2020;64(1-2-3):239-246. doi: 10.1387/ijdb.190148mm. Int J Dev Biol. 2020. PMID: 32659012 Review.
-
Neuronal Calcium Signaling in Metabolic Regulation and Adaptation to Nutrient Stress.Front Neural Circuits. 2018 Apr 5;12:25. doi: 10.3389/fncir.2018.00025. eCollection 2018. Front Neural Circuits. 2018. PMID: 29674958 Free PMC article. Review.
Cited by
-
Nutrient responding peptide hormone CCHamide-2 consolidates appetitive memory.Front Behav Neurosci. 2022 Oct 19;16:986064. doi: 10.3389/fnbeh.2022.986064. eCollection 2022. Front Behav Neurosci. 2022. PMID: 36338876 Free PMC article.
-
Neural pathways in nutrient sensing and insulin signaling.Front Physiol. 2022 Nov 11;13:1002183. doi: 10.3389/fphys.2022.1002183. eCollection 2022. Front Physiol. 2022. PMID: 36439265 Free PMC article. Review.
-
Starvation selection reduces and delays larval ecdysone production and signaling.J Exp Biol. 2023 Sep 15;226(18):jeb246144. doi: 10.1242/jeb.246144. Epub 2023 Sep 29. J Exp Biol. 2023. PMID: 37671530 Free PMC article.
-
ER-Ca2+ sensor STIM regulates neuropeptides required for development under nutrient restriction in Drosophila.PLoS One. 2019 Jul 11;14(7):e0219719. doi: 10.1371/journal.pone.0219719. eCollection 2019. PLoS One. 2019. PMID: 31295329 Free PMC article.
-
Growth regulation by amino acid transporters in Drosophila larvae.Cell Mol Life Sci. 2020 Nov;77(21):4289-4297. doi: 10.1007/s00018-020-03535-6. Epub 2020 May 1. Cell Mol Life Sci. 2020. PMID: 32358623 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
