mTOR inhibitors lower an intrinsic barrier to virus infection mediated by IFITM3
- PMID: 30301809
- PMCID: PMC6205447
- DOI: 10.1073/pnas.1811892115
mTOR inhibitors lower an intrinsic barrier to virus infection mediated by IFITM3
Abstract
Rapamycin and its derivatives are specific inhibitors of mammalian target of rapamycin (mTOR) kinase and, as a result, are well-established immunosuppressants and antitumorigenic agents. Additionally, this class of drug promotes gene delivery by facilitating lentiviral vector entry into cells, revealing its potential to improve gene therapy efforts. However, the precise mechanism was unknown. Here, we report that mTOR inhibitor treatment results in down-regulation of the IFN-induced transmembrane (IFITM) proteins. IFITM proteins, especially IFITM3, are potent inhibitors of virus-cell fusion and are broadly active against a range of pathogenic viruses. We found that the effect of rapamycin treatment on lentiviral transduction is diminished upon IFITM silencing or knockout in primary and transformed cells, and the extent of transduction enhancement depends on basal expression of IFITM proteins, with a major contribution from IFITM3. The effect of rapamycin treatment on IFITM3 manifests at the level of protein, but not mRNA, and is selective, as many other endosome-associated transmembrane proteins are unaffected. Rapamycin-mediated degradation of IFITM3 requires endosomal trafficking, ubiquitination, endosomal sorting complex required for transport (ESCRT) machinery, and lysosomal acidification. Since IFITM proteins exhibit broad antiviral activity, we show that mTOR inhibition also promotes infection by another IFITM-sensitive virus, Influenza A virus, but not infection by Sendai virus, which is IFITM-resistant. Our results identify the molecular basis by which mTOR inhibitors enhance virus entry into cells and reveal a previously unrecognized immunosuppressive feature of these clinically important drugs. In addition, this study uncovers a functional convergence between the mTOR pathway and IFITM proteins at endolysosomal membranes.
Keywords: IFITM; endosome; fusion; interferon; virus.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
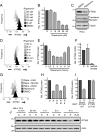
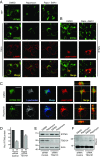
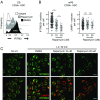

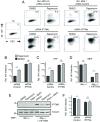
Similar articles
-
Interferon-induced transmembrane protein 3 blocks fusion of sensitive but not resistant viruses by partitioning into virus-carrying endosomes.PLoS Pathog. 2019 Jan 14;15(1):e1007532. doi: 10.1371/journal.ppat.1007532. eCollection 2019 Jan. PLoS Pathog. 2019. PMID: 30640957 Free PMC article.
-
Duck Interferon-Inducible Transmembrane Protein 3 Mediates Restriction of Influenza Viruses.J Virol. 2015 Oct 14;90(1):103-16. doi: 10.1128/JVI.01593-15. Print 2016 Jan 1. J Virol. 2015. PMID: 26468537 Free PMC article.
-
IFITM Proteins That Restrict the Early Stages of Respiratory Virus Infection Do Not Influence Late-Stage Replication.J Virol. 2021 Sep 27;95(20):e0083721. doi: 10.1128/JVI.00837-21. Epub 2021 Jul 28. J Virol. 2021. PMID: 34319159 Free PMC article.
-
Discovery and Characterization of IFITM S-Palmitoylation.Viruses. 2023 Nov 28;15(12):2329. doi: 10.3390/v15122329. Viruses. 2023. PMID: 38140570 Free PMC article. Review.
-
More than meets the I: the diverse antiviral and cellular functions of interferon-induced transmembrane proteins.Retrovirology. 2017 Nov 21;14(1):53. doi: 10.1186/s12977-017-0377-y. Retrovirology. 2017. PMID: 29162141 Free PMC article. Review.
Cited by
-
Site-Specific Photo-Crosslinking Proteomics Reveal Regulation of IFITM3 Trafficking and Turnover by VCP/p97 ATPase.Cell Chem Biol. 2020 May 21;27(5):571-585.e6. doi: 10.1016/j.chembiol.2020.03.004. Epub 2020 Apr 2. Cell Chem Biol. 2020. PMID: 32243810 Free PMC article.
-
IFITM protein regulation and functions: Far beyond the fight against viruses.Front Immunol. 2022 Nov 18;13:1042368. doi: 10.3389/fimmu.2022.1042368. eCollection 2022. Front Immunol. 2022. PMID: 36466909 Free PMC article. Review.
-
p53 promotes ZDHHC1-mediated IFITM3 palmitoylation to inhibit Japanese encephalitis virus replication.PLoS Pathog. 2020 Oct 27;16(10):e1009035. doi: 10.1371/journal.ppat.1009035. eCollection 2020 Oct. PLoS Pathog. 2020. Retraction in: PLoS Pathog. 2024 Nov 8;20(11):e1012698. doi: 10.1371/journal.ppat.1012698. PMID: 33108395 Free PMC article. Retracted.
-
Assessing the Impact of Cyclosporin A on Lentiviral Transduction and Preservation of Human Hematopoietic Stem Cells in Clinically Relevant Ex Vivo Gene Therapy Settings.Hum Gene Ther. 2019 Sep;30(9):1133-1146. doi: 10.1089/hum.2019.016. Epub 2019 May 24. Hum Gene Ther. 2019. PMID: 31037976 Free PMC article.
-
The Binary Model of Chronic Diseases Applied to COVID-19.Front Immunol. 2021 Sep 3;12:716084. doi: 10.3389/fimmu.2021.716084. eCollection 2021. Front Immunol. 2021. PMID: 34539649 Free PMC article.
References
-
- Alberts B, et al. Molecular Biology of the Cell. 6th Ed Garland Science; New York: 2014.
-
- Luzio JP, Pryor PR, Bright NA. Lysosomes: Fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

