Microglia innately develop within cerebral organoids
- PMID: 30301888
- PMCID: PMC6177485
- DOI: 10.1038/s41467-018-06684-2
Microglia innately develop within cerebral organoids
Abstract
Cerebral organoids are 3D stem cell-derived models that can be utilized to study the human brain. The current consensus is that cerebral organoids consist of cells derived from the neuroectodermal lineage. This limits their value and applicability, as mesodermal-derived microglia are important players in neural development and disease. Remarkably, here we show that microglia can innately develop within a cerebral organoid model and display their characteristic ramified morphology. The transcriptome and response to inflammatory stimulation of these organoid-grown microglia closely mimic the transcriptome and response of adult microglia acutely isolated from post mortem human brain tissue. In addition, organoid-grown microglia mediate phagocytosis and synaptic material is detected inside them. In all, our study characterizes a microglia-containing organoid model that represents a valuable tool for studying the interplay between microglia, macroglia, and neurons in human brain development and disease.
Conflict of interest statement
L.H.B. declares the following competing financial interests: travel grants and consultancy fees at Baxter International, and is a member of the scientific advisory board at Biogen Idec, Cytokinetics, and no other competing interests. All other authors declare no competing interests.
Figures
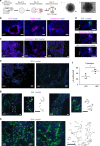
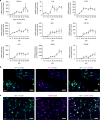

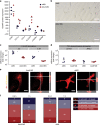

Comment in
-
Findings of Research Misconduct.Fed Regist. 2020 Aug 14;85(158):49661-49662. Fed Regist. 2020. PMID: 32831428 Free PMC article. No abstract available.
References
-
- Stevens Beth, Allen Nicola J., Vazquez Luis E., Howell Gareth R., Christopherson Karen S., Nouri Navid, Micheva Kristina D., Mehalow Adrienne K., Huberman Andrew D., Stafford Benjamin, Sher Alexander, Litke Alan M., Lambris John D., Smith Stephen J., John Simon W.M., Barres Ben A. The Classical Complement Cascade Mediates CNS Synapse Elimination. Cell. 2007;131(6):1164–1178. doi: 10.1016/j.cell.2007.10.036. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

