Functional analysis of polyketide synthase genes in the biocontrol fungus Clonostachys rosea
- PMID: 30301915
- PMCID: PMC6177402
- DOI: 10.1038/s41598-018-33391-1
Functional analysis of polyketide synthase genes in the biocontrol fungus Clonostachys rosea
Abstract
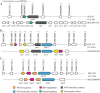
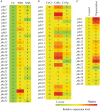
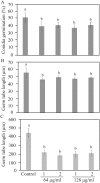
Clonostachys rosea is a mycoparasitic fungus used for biological control of plant diseases. Its genome contains 31 genes putatively encoding for polyketide synthases (PKSs), 75% of which are arranged in biosynthetic gene clusters. Gene expression analysis during C. rosea interactions with the fungal plant pathogens Botrytis cinerea and Fusarium graminearum showed common and species-specific induction of PKS genes. Our data showed a culture media dependent correlation between PKS gene expression and degree of antagonism in C. rosea. The pks22 and pks29 genes were highly induced during fungal-fungal interactions but not during pigmentation, and gene deletion studies revealed that PKS29 was required for full antagonism against B. cinerea, and for biocontrol of fusarium foot rot on barley. Metabolite analysis revealed that Δpks29 strains has a 50% reduced production (P = 0.001) of an unknown polyketide with molecular formula C15H28O3, while Δpks22 strains lost the ability to produce four previously unknown polyketides named Clonorosein A-D. Clonorosein A and B were purified, their structures determined, and showed strong antifungal activity against B. cinerea and F. graminearum. These results show that PKS22 is required for production of antifungal polyketide Clonorosein A-D, and demonstrate the role of PKS29 in antagonism and biocontrol of fungal plant diseases.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

