Pleiotropy of the Drosophila melanogaster foraging gene on larval feeding-related traits
- PMID: 30303018
- PMCID: PMC6309726
- DOI: 10.1080/01677063.2018.1500572
Pleiotropy of the Drosophila melanogaster foraging gene on larval feeding-related traits
Abstract
Little is known about the molecular underpinning of behavioral pleiotropy. The Drosophila melanogaster foraging gene is highly pleiotropic, affecting many independent larval and adult phenotypes. Included in foraging's multiple phenotypes are larval foraging path length, triglyceride levels, and food intake. foraging has a complex structure with four promoters and 21 transcripts that encode nine protein isoforms of a cGMP dependent protein kinase (PKG). We examined if foraging's complex molecular structure underlies the behavioral pleiotropy associated with this gene. Using a promotor analysis strategy, we cloned DNA fragments upstream of each of foraging's transcription start sites and generated four separate forpr-Gal4s. Supporting our hypothesis of modular function, they had discrete, restricted expression patterns throughout the larva. In the CNS, forpr1-Gal4 and forpr4-Gal4 were expressed in neurons while forpr2-Gal4 and forpr3-Gal4 were expressed in glia cells. In the gastric system, forpr1-Gal4 and forpr3-Gal4 were expressed in enteroendocrine cells of the midgut while forpr2-Gal4 was expressed in the stem cells of the midgut. forpr3-Gal4 was expressed in the midgut enterocytes, and midgut and hindgut visceral muscle. forpr4-Gal4's gastric system expression was restricted to the hindgut. We also found promoter specific expression in the larval fat body, salivary glands, and body muscle. The modularity of foraging's molecular structure was also apparent in the phenotypic rescues. We rescued larval path length, triglyceride levels (bordered on significance), and food intake of for0 null larvae using different forpr-Gal4s to drive UAS-forcDNA. In a foraging null genetic background, forpr1-Gal4 was the only promoter driven Gal4 to rescue larval path length, forpr3-Gal4 altered triglyceride levels, and forpr4-Gal4 rescued food intake. Our results refine the spatial expression responsible for foraging's associated phenotypes, as well as the sub-regions of the locus responsible for their expression. foraging's pleiotropy arises at least in part from the individual contributions of its four promoters.
Keywords: Drosophila melanogaster; foraging gene; larval phenotypes; multiple promoter analysis; pleiotropy.
Conflict of interest statement
Disclosure of interest:
The authors report no conflict of interest.
Figures


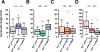



References
-
- Andersson L, & Georges M (2004). Domestic-animal genomics: deciphering the genetics of complex traits. Nature Reviews. Genetics, 5(3), 202–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
