Global genetic insight contributed by consanguineous Pakistani families segregating hearing loss
- PMID: 30303587
- PMCID: PMC6296877
- DOI: 10.1002/humu.23666
Global genetic insight contributed by consanguineous Pakistani families segregating hearing loss
Abstract
Consanguineous Pakistani pedigrees segregating deafness have contributed decisively to the discovery of 31 of the 68 genes associated with nonsyndromic autosomal recessive hearing loss (HL) worldwide. In this study, we utilized genome-wide genotyping, Sanger and exome sequencing to identify 163 DNA variants in 41 previously reported HL genes segregating in 321 Pakistani families. Of these, 70 (42.9%) variants identified in 29 genes are novel. As expected from genetic studies of disorders segregating in consanguineous families, the majority of affected individuals (94.4%) are homozygous for HL-associated variants, with the other variants being compound heterozygotes. The five most common HL genes in the Pakistani population are SLC26A4, MYO7A, GJB2, CIB2 and HGF, respectively. Our study provides a profile of the genetic etiology of HL in Pakistani families, which will allow for the development of more efficient genetic diagnostic tools, aid in accurate genetic counseling, and guide application of future gene-based therapies. These findings are also valuable in interpreting pathogenicity of variants that are potentially associated with HL in individuals of all ancestries. The Pakistani population, and its infrastructure for studying human genetics, will continue to be valuable to gene discovery for HL and other inherited disorders.
Keywords: DFNB; Pakistan; allelic heterogeneity; autosomal recessive hearing loss; deafness; genetic spectrum; pathogenic variant.
© 2018 Wiley Periodicals, Inc.
Conflict of interest statement
Disclosure statement: All authors declare no conflicts of interest.
Figures

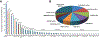
References
-
- Anjum S, Azhar A, Tariq M, Baig S, HJ. B, Qayyum M, … Raja G. (2014). GJB2 Gene Mutations Causing Hearing Loss in Consanguineous Pakistani Families. Pak J Life Soc Sci(12), 126–131.
Publication types
MeSH terms
Grants and funding
- U54 HG006493/HG/NHGRI NIH HHS/United States
- R01 DC003594/GF/NIH HHS/United States
- R01 DC011651/DC/NIDCD NIH HHS/United States
- R01 DC012564/GF/NIH HHS/United States
- R01 DC012564/DC/NIDCD NIH HHS/United States
- R01 DC011651/GF/NIH HHS/United States
- R56 DC011803/DC/NIDCD NIH HHS/United States
- R01 DC000086/DC/NIDCD NIH HHS/United States
- HG006493/Higher Education Commission of Pakistan/International
- DC000039-20/DC/NIDCD NIH HHS/United States
- DC000086/DC/NIDCD NIH HHS/United States
- UM1 HG006493/HG/NHGRI NIH HHS/United States
- Z01 DC000039/ImNIH/Intramural NIH HHS/United States
- R01 DC016295/DC/NIDCD NIH HHS/United States
- R56 DC011803/GF/NIH HHS/United States
- R01 DC003594/DC/NIDCD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

