Complete genome sequences of two strains of Treponema pallidum subsp. pertenue from Indonesia: Modular structure of several treponemal genes
- PMID: 30303967
- PMCID: PMC6197692
- DOI: 10.1371/journal.pntd.0006867
Complete genome sequences of two strains of Treponema pallidum subsp. pertenue from Indonesia: Modular structure of several treponemal genes
Abstract
Background: Treponema pallidum subsp. pertenue (TPE) is the causative agent of yaws, a multistage disease endemic in tropical regions in Africa, Asia, Oceania, and South America. To date, seven TPE strains have been completely sequenced and analyzed including five TPE strains of human origin (CDC-2, CDC 2575, Gauthier, Ghana-051, and Samoa D) and two TPE strains isolated from the baboons (Fribourg-Blanc and LMNP-1). This study revealed the complete genome sequences of two TPE strains, Kampung Dalan K363 and Sei Geringging K403, isolated in 1990 from villages in the Pariaman region of Sumatra, Indonesia and compared these genome sequences with other known TPE genomes.
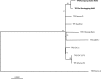
Methodology/principal findings: The genomes were determined using the pooled segment genome sequencing method combined with the Illumina sequencing platform resulting in an average coverage depth of 1,021x and 644x for the TPE Kampung Dalan K363 and TPE Sei Geringging K403 genomes, respectively. Both Indonesian TPE strains were genetically related to each other and were more distantly related to other, previously characterized TPE strains. The modular character of several genes, including TP0136 and TP0858 gene orthologs, was identified by analysis of the corresponding sequences. To systematically detect genes potentially having a modular genetic structure, we performed a whole genome analysis-of-occurrence of direct or inverted repeats of 17 or more nucleotides in length. Besides in tpr genes, a frequent presence of repeats was found in the genetic regions spanning TP0126-TP0136, TP0856-TP0858, and TP0896 genes.
Conclusions/significance: Comparisons of genome sequences of TPE Kampung Dalan K363 and Sei Geringging K403 with other TPE strains revealed a modular structure of several genomic loci including the TP0136, TP0856, and TP0858 genes. Diversification of TPE genomes appears to be facilitated by intra-strain genome recombination events.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Complete genome sequences of two strains of Treponema pallidum subsp. pertenue from Ghana, Africa: Identical genome sequences in samples isolated more than 7 years apart.PLoS Negl Trop Dis. 2017 Sep 8;11(9):e0005894. doi: 10.1371/journal.pntd.0005894. eCollection 2017 Sep. PLoS Negl Trop Dis. 2017. PMID: 28886021 Free PMC article.
-
Whole genome sequence of the Treponema Fribourg-Blanc: unspecified simian isolate is highly similar to the yaws subspecies.PLoS Negl Trop Dis. 2013 Apr 18;7(4):e2172. doi: 10.1371/journal.pntd.0002172. Print 2013. PLoS Negl Trop Dis. 2013. PMID: 23638193 Free PMC article.
-
Whole genome sequences of three Treponema pallidum ssp. pertenue strains: yaws and syphilis treponemes differ in less than 0.2% of the genome sequence.PLoS Negl Trop Dis. 2012 Jan;6(1):e1471. doi: 10.1371/journal.pntd.0001471. Epub 2012 Jan 24. PLoS Negl Trop Dis. 2012. PMID: 22292095 Free PMC article.
-
Genetics of human and animal uncultivable treponemal pathogens.Infect Genet Evol. 2018 Jul;61:92-107. doi: 10.1016/j.meegid.2018.03.015. Epub 2018 Mar 22. Infect Genet Evol. 2018. PMID: 29578082 Review.
-
Genetic diversity in Treponema pallidum: implications for pathogenesis, evolution and molecular diagnostics of syphilis and yaws.Infect Genet Evol. 2012 Mar;12(2):191-202. doi: 10.1016/j.meegid.2011.12.001. Epub 2011 Dec 15. Infect Genet Evol. 2012. PMID: 22198325 Free PMC article. Review.
Cited by
-
MLST typing of Treponema pallidum subsp. pallidum in the Czech Republic during 2004-2017: Clinical isolates belonged to 25 allelic profiles and harbored 8 novel allelic variants.PLoS One. 2019 May 31;14(5):e0217611. doi: 10.1371/journal.pone.0217611. eCollection 2019. PLoS One. 2019. PMID: 31150464 Free PMC article.
-
Identification of positively selected genes in human pathogenic treponemes: Syphilis-, yaws-, and bejel-causing strains differ in sets of genes showing adaptive evolution.PLoS Negl Trop Dis. 2019 Jun 19;13(6):e0007463. doi: 10.1371/journal.pntd.0007463. eCollection 2019 Jun. PLoS Negl Trop Dis. 2019. PMID: 31216284 Free PMC article.
-
Evolutionary Processes in the Emergence and Recent Spread of the Syphilis Agent, Treponema pallidum.Mol Biol Evol. 2022 Jan 7;39(1):msab318. doi: 10.1093/molbev/msab318. Mol Biol Evol. 2022. PMID: 34791386 Free PMC article.
-
Directly Sequenced Genomes of Contemporary Strains of Syphilis Reveal Recombination-Driven Diversity in Genes Encoding Predicted Surface-Exposed Antigens.Front Microbiol. 2019 Jul 31;10:1691. doi: 10.3389/fmicb.2019.01691. eCollection 2019. Front Microbiol. 2019. PMID: 31417509 Free PMC article.
-
Insights into Treponema pallidum genomics from modern and ancient genomes using a novel mapping strategy.BMC Biol. 2025 Jan 8;23(1):7. doi: 10.1186/s12915-024-02108-4. BMC Biol. 2025. PMID: 39780098 Free PMC article.
References
-
- Mikalová L, Strouhal M, Oppelt J, Grange PA, Janier M, Benhaddou N, et al. Human Treponema pallidum 11q/j isolate belongs to subsp. endemicum but contains two loci with a sequence in TP0548 and TP0488 similar to subsp. pertenue and subsp. pallidum, respectively. PLoS Negl Trop Dis. 2017;11: e0005434 10.1371/journal.pntd.0005434 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

