Regulation of muscle atrophy-related genes by the opposing transcriptional activities of ZEB1/CtBP and FOXO3
- PMID: 30304480
- PMCID: PMC6237734
- DOI: 10.1093/nar/gky835
Regulation of muscle atrophy-related genes by the opposing transcriptional activities of ZEB1/CtBP and FOXO3
Abstract
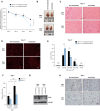
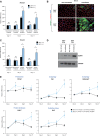
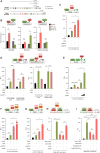
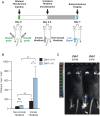
Multiple physiopathological and clinical conditions trigger skeletal muscle atrophy through the induction of a group of proteins (atrogenes) that includes components of the ubiquitin-proteasome and autophagy-lysosomal systems. Atrogenes are induced by FOXO transcription factors, but their regulation is still not fully understood. Here, we showed that the transcription factor ZEB1, best known for promoting tumor progression, inhibits muscle atrophy and atrogene expression by antagonizing FOXO3-mediated induction of atrogenes. Compared to wild-type counterparts, hindlimb immobilization in Zeb1-deficient mice resulted in enhanced muscle atrophy and higher expression of a number of atrogenes, including Atrogin-1/Fbxo32, MuRF1/Trim63, Ctsl, 4ebp1, Gabarapl1, Psma1 and Nrf2. Likewise, in the C2C12 myogenic cell model, ZEB1 knockdown augmented both myotube diameter reduction and atrogene upregulation in response to nutrient deprivation. Mechanistically, ZEB1 directly represses in vitro and in vivo Fbxo32 and Trim63 promoter transcription in a stage-dependent manner and in a reverse pattern with MYOD1. ZEB1 bound to the Fbxo32 promoter in undifferentiated myoblasts and atrophic myotubes, but not in non-atrophic myotubes, where it is displaced by MYOD1. ZEB1 repressed both promoters through CtBP-mediated inhibition of FOXO3 transcriptional activity. These results set ZEB1 as a new target in therapeutic approaches to clinical conditions causing muscle mass loss.
Figures


References
-
- Cohen S., Nathan J.A., Goldberg A.L.. Muscle wasting in disease: molecular mechanisms and promising therapies. Nat. Rev. Drug Discov. 2015; 14:58–74. - PubMed
-
- Schiaffino S., Dyar K.A., Ciciliot S., Blaauw B., Sandri M.. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013; 280:4294–4314. - PubMed
-
- Bodine S.C., Latres E., Baumhueter S., Lai V.K.M., Nunez L., Clarke B.A., Poueymirou W.T., Panaro F.J., Na E., Dharmarajan K. et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001; 294:1704–1708. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

