Whole Blood Gene Expression Reveals Specific Transcriptome Changes in Neonatal Encephalopathy
- PMID: 30304723
- PMCID: PMC6425817
- DOI: 10.1159/000492420
Whole Blood Gene Expression Reveals Specific Transcriptome Changes in Neonatal Encephalopathy
Abstract
Background: Variable responses to hypothermic neuroprotection are related to the clinical heterogeneity of encephalopathic babies; hence better disease stratification may facilitate the development of individualized neuroprotective therapies.
Objectives: We examined if whole blood gene expression analysis can identify specific transcriptome profiles in neonatal encephalopathy.
Material and methods: We performed next-generation sequencing on whole blood RNA from 12 babies with neonatal encephalopathy and 6 time-matched healthy term babies. Genes significantly differentially expressed between encephalopathic and control babies were identified. This set of genes was then compared to the host RNA response in septic neonates and subjected to pathway analysis.
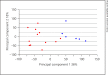
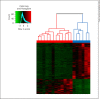
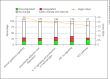
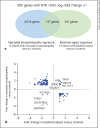
Results: We identified 950 statistically significant genes discriminating perfectly between healthy controls and neonatal encephalopathy. The major pathways in neonatal encephalopathy were axonal guidance signaling (p = 0.0009), granulocyte adhesion and diapedesis (p = 0.003), IL-12 signaling and production in macrophages (p = 0.003), and hypoxia-inducible factor 1α signaling (p = 0.004). There were only 137 genes in common between neonatal encephalopathy and bacterial sepsis sets.
Conclusion: Babies with neonatal encephalopathy have striking differences in gene expression profiles compared with healthy control and septic babies. Gene expression profiles may be useful for disease stratification and for developing personalized neuroprotective therapies.
Keywords: Biomarkers; Brain injury; Gene expression; Neonatal encephalopathy.
The Author(s). Published by S. Karger AG, Basel.
Figures

References
-
- Whitelaw A, Thoresen M. Animal research has been essential to saving babies' lives. BMJ. 2014 Jun;348(jun24 5):g4174. - PubMed
-
- Osredkar D, Thoresen M, Maes E, Flatebø T, Elstad M, Sabir H. Hypothermia is not neuroprotective after infection-sensitized neonatal hypoxic-ischemic brain injury. Resuscitation. 2014 Apr;85((4)):567–72. - PubMed

