Exploring the Remote Ties between Helitron Transposases and Other Rolling-Circle Replication Proteins
- PMID: 30304800
- PMCID: PMC6213432
- DOI: 10.3390/ijms19103079
Exploring the Remote Ties between Helitron Transposases and Other Rolling-Circle Replication Proteins
Abstract
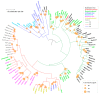
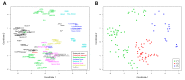
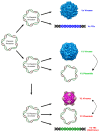
Rolling-circle replication (RCR) elements constitute a diverse group that includes viruses, plasmids, and transposons, present in hosts from all domains of life. Eukaryotic RCR transposons, also known as Helitrons, are found in species from all eukaryotic kingdoms, sometimes representing a large portion of their genomes. Despite the impact of Helitrons on their hosts, knowledge about their relationship with other RCR elements is still elusive. Here, we compared the endonuclease domain sequence of Helitron transposases with the corresponding region from RCR proteins found in a wide variety of mobile genetic elements. To do that, we used a stepwise alignment approach followed by phylogenetic and multidimensional scaling analyses. Although it has been suggested that Helitrons might have originated from prokaryotic transposons or eukaryotic viruses, our results indicate that Helitron transposases share more similarities with proteins from prokaryotic viruses and plasmids instead. We also provide evidence for the division of RCR endonucleases into three groups (Y1, Y2, and Yx), covering the whole diversity of this protein family. Together, these results point to prokaryotic elements as the likely closest ancestors of eukaryotic RCR transposons, and further demonstrate the fluidity that characterizes the boundaries separating viruses, plasmids, and transposons.
Keywords: Helitron; mobile genetic element; rolling-circle replication; viral evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Replitrons: A major group of eukaryotic transposons encoding HUH endonuclease.Proc Natl Acad Sci U S A. 2023 Jun 20;120(25):e2301424120. doi: 10.1073/pnas.2301424120. Epub 2023 Jun 12. Proc Natl Acad Sci U S A. 2023. PMID: 37307447 Free PMC article.
-
Pif1 Helicases and the Evidence for a Prokaryotic Origin of Helitrons.Mol Biol Evol. 2022 Jan 7;39(1):msab334. doi: 10.1093/molbev/msab334. Mol Biol Evol. 2022. PMID: 34850089 Free PMC article.
-
Rolling-circle amplification of centromeric Helitrons in plant genomes.Plant J. 2016 Dec;88(6):1038-1045. doi: 10.1111/tpj.13314. Epub 2016 Oct 25. Plant J. 2016. PMID: 27553634
-
Helitrons on a roll: eukaryotic rolling-circle transposons.Trends Genet. 2007 Oct;23(10):521-9. doi: 10.1016/j.tig.2007.08.004. Epub 2007 Sep 11. Trends Genet. 2007. PMID: 17850916 Review.
-
Helitrons: genomic parasites that generate developmental novelties.Trends Genet. 2024 May;40(5):437-448. doi: 10.1016/j.tig.2024.02.002. Epub 2024 Feb 29. Trends Genet. 2024. PMID: 38429198 Review.
Cited by
-
Discovery of numerous novel Helitron-like elements in eukaryote genomes using HELIANO.Nucleic Acids Res. 2024 Sep 23;52(17):e79. doi: 10.1093/nar/gkae679. Nucleic Acids Res. 2024. PMID: 39119924 Free PMC article.
-
Replitrons: A major group of eukaryotic transposons encoding HUH endonuclease.Proc Natl Acad Sci U S A. 2023 Jun 20;120(25):e2301424120. doi: 10.1073/pnas.2301424120. Epub 2023 Jun 12. Proc Natl Acad Sci U S A. 2023. PMID: 37307447 Free PMC article.
-
Pif1 Helicases and the Evidence for a Prokaryotic Origin of Helitrons.Mol Biol Evol. 2022 Jan 7;39(1):msab334. doi: 10.1093/molbev/msab334. Mol Biol Evol. 2022. PMID: 34850089 Free PMC article.
-
The large bat Helitron DNA transposase forms a compact monomeric assembly that buries and protects its covalently bound 5'-transposon end.Mol Cell. 2021 Oct 21;81(20):4271-4286.e4. doi: 10.1016/j.molcel.2021.07.028. Epub 2021 Aug 16. Mol Cell. 2021. PMID: 34403695 Free PMC article.
-
A Field Guide to Eukaryotic Transposable Elements.Annu Rev Genet. 2020 Nov 23;54:539-561. doi: 10.1146/annurev-genet-040620-022145. Epub 2020 Sep 21. Annu Rev Genet. 2020. PMID: 32955944 Free PMC article. Review.
References
-
- Thomas J., Pritham E.J. Helitrons, the Eukaryotic Rolling-Circle Transposable Elements in Mobile DNA III. 3rd ed. ASM Press; Washington, DC, USA: 2015. pp. 893–926. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

