Cytoplasmic p21 Mediates 5-Fluorouracil Resistance by Inhibiting Pro-Apoptotic Chk2
- PMID: 30304835
- PMCID: PMC6210175
- DOI: 10.3390/cancers10100373
Cytoplasmic p21 Mediates 5-Fluorouracil Resistance by Inhibiting Pro-Apoptotic Chk2
Abstract
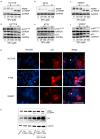
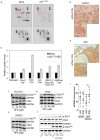
The oncogenic cytoplasmic p21 contributes to cancer aggressiveness and chemotherapeutic failure. However, the molecular mechanisms remain obscure. Here, we show for the first time that cytoplasmic p21 mediates 5-Fluorouracil (5FU) resistance by shuttling p-Chk2 out of the nucleus to protect the tumor cells from its pro-apoptotic functions. We observed that cytoplasmic p21 levels were up-regulated in 5FU-resistant colorectal cancer cells in vitro and the in vivo Chorioallantoic membrane (CAM) model. Kinase array analysis revealed that p-Chk2 is a key target of cytoplasmic p21. Importantly, cytoplasmic form of p21 mediated by p21T145D transfection diminished p-Chk2-mediated activation of E2F1 and apoptosis induction. Co-immunoprecipitation, immunofluorescence, and proximity ligation assay showed that p21 forms a complex with p-Chk2 under 5FU exposure. Using in silico computer modeling, we suggest that the p21/p-Chk2 interaction hindered the nuclear localization signal of p-Chk2, and therefore, the complex is exported out of the nucleus. These findings unravel a novel mechanism regarding an oncogenic role of p21 in regulation of resistance to 5FU-based chemotherapy. We suggest a possible value of cytoplasmic p21 as a prognosis marker and a therapeutic target in colorectal cancer patients.
Keywords: 5-fluorouracil resistance; Chk2; colorectal cancer; cytoplasmic p21; p21; protein interaction.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







References
-
- Efficacy of adjuvant fluorouracil and folinic acid in colon cancer International Multicentre Pooled Analysis of Colon Cancer Trials (IMPACT) investigators. Lancet. 1995;345:939–944. - PubMed
-
- Zhang S., Hu B., You Y., Yang Z., Liu L., Tang H., Bao W., Guan Y., Shen X. Sorting nexin 10 acts as a tumor suppressor in tumorigenesis and progression of colorectal cancer through regulating chaperone mediated autophagy degradation of p21(Cip1/WAF1) Cancer Lett. 2018;419:116–127. doi: 10.1016/j.canlet.2018.01.045. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

