Epigenetic Modifiers in Myeloid Malignancies: The Role of Histone Deacetylase Inhibitors
- PMID: 30304859
- PMCID: PMC6212943
- DOI: 10.3390/ijms19103091
Epigenetic Modifiers in Myeloid Malignancies: The Role of Histone Deacetylase Inhibitors
Abstract
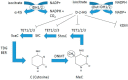
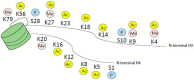
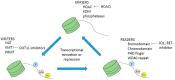
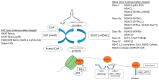
Myeloid hematological malignancies are clonal bone marrow neoplasms, comprising of acute myeloid leukemia (AML), the myelodysplastic syndromes (MDS), chronic myelomonocytic leukemia (CMML), the myeloproliferative neoplasms (MPN) and systemic mastocytosis (SM). The field of epigenetic regulation of normal and malignant hematopoiesis is rapidly growing. In recent years, heterozygous somatic mutations in genes encoding epigenetic regulators have been found in all subtypes of myeloid malignancies, supporting the rationale for treatment with epigenetic modifiers. Histone deacetylase inhibitors (HDACi) are epigenetic modifiers that, in vitro, have been shown to induce growth arrest, apoptotic or autophagic cell death, and terminal differentiation of myeloid tumor cells. These effects were observed both at the bulk tumor level and in the most immature CD34⁺38- cell compartments containing the leukemic stem cells. Thus, there is a strong rationale supporting HDACi therapy in myeloid malignancies. However, despite initial promising results in phase I trials, HDACi in monotherapy as well as in combination with other drugs, have failed to improve responses or survival. This review provides an overview of the rationale for HDACi in myeloid malignancies, clinical results and speculations on why clinical trials have thus far not met the expectations, and how this may be improved in the future.
Keywords: acute myeloid leukemia; chronic myelomonocytic leukemia; myelodysplastic syndromes; myeloid mutations; systemic mastocytosis; treatment.
Conflict of interest statement
The author declares that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Figures
References
-
- Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. - DOI - PubMed
-
- Daskalakis M., Nguyen T.T., Nguyen C., Guldberg P., Kohler G., Wijermans P., Jones P.A., Lubbert M. Demethylation of a hypermethylated P15/INK4B gene in patients with myelodysplastic syndrome by 5-Aza-2′-deoxycytidine (decitabine) treatment. Blood. 2002;100:2957–2964. doi: 10.1182/blood.V100.8.2957. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

