Local activation of mammalian separase in interphase promotes double-strand break repair and prevents oncogenic transformation
- PMID: 30305303
- PMCID: PMC6236338
- DOI: 10.15252/embj.201899184
Local activation of mammalian separase in interphase promotes double-strand break repair and prevents oncogenic transformation
Abstract
Separase halves eukaryotic chromosomes in M-phase by cleaving cohesin complexes holding sister chromatids together. Whether this essential protease functions also in interphase and/or impacts carcinogenesis remains largely unknown. Here, we show that mammalian separase is recruited to DNA double-strand breaks (DSBs) where it is activated to locally cleave cohesin and facilitate homology-directed repair (HDR). Inactivating phosphorylation of its NES, arginine methylation of its RG-repeats, and sumoylation redirect separase from the cytosol to DSBs. In vitro assays suggest that DNA damage response-relevant ATM, PRMT1, and Mms21 represent the corresponding kinase, methyltransferase, and SUMO ligase, respectively. SEPARASE heterozygosity not only debilitates HDR but also predisposes primary embryonic fibroblasts to neoplasia and mice to chemically induced skin cancer. Thus, tethering of separase to DSBs and confined cohesin cleavage promote DSB repair in G2 cells. Importantly, this conserved interphase function of separase protects mammalian cells from oncogenic transformation.
Keywords: DNA double‐strand breaks; cohesin; homology‐directed repair; posttranslational modifications; separase.
© 2018 The Authors.
Figures
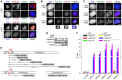
A system for induced introduction of site‐specific DSBs. Transgenic HEK293 cells constitutively expressing FLAG‐tagged AsiSI‐ER were treated with OHT or carrier solvent and then analyzed by IFM as indicated. Scale bar = 5 μm.
Transgenic HEK293 cells treated with Dox to induce expression of Myc‐separase‐WT and/or with OHT to induce nuclear accumulation of ER‐AsiSI and DSBs were analyzed by IFM as indicated. Scale bar = 5 μm.
γH2AX‐ and separase‐positive foci formation in response to DNA damage by DRB. Prior to their analysis by IFM, transgenic HEK293 cells in G2‐phase were Dox‐ and/or DRB‐treated to induce the expression of Myc‐separase and/or inflict DSBs, respectively. Scale bar = 5 μm.
Western blot analysis of experiment shown in Fig 1A.
Position and sizes of PCR fragments from the ChIP–multiplex PCR and ChIP‐qPCR experiments. Schematic is not drawn to scale.
Transgenic HEK293 cells supplemented with Dox to induce expression of Myc‐separase‐WT were mock‐ or OHT‐treated in G2‐phase and then subjected to ChIP‐qPCR. Shown are averages (bars) of three independent experiments (dots).

- A
Following transfection of given siRNAs and synchronization in G2‐phase, Hek293 cells were DRB‐ or mock‐treated and then subjected to IFM using the indicated antibodies. Lower panels display a threefold magnification of the boxed area shown above. Scales bars correspond to 5 and 1 μm, respectively. See Fig EV1D for corresponding immunoblot.
- B
HEK293 cells were thymidine‐arrested for 20 h, mock‐ or OHT‐treated to induce DSBs by nuclear accumulation of ER‐AsiSI, and then subjected to ChIP–multiplex PCR.
- C
ChIP samples from (B) were analyzed by qPCR. Shown are averages (bars) of three independent experiments (dots).
- D, E
Separase interacts with γH2AX in DSB‐containing G2 but not G1 cells. Transgenic HEK293 cells treated with Dox to induce expression of Myc‐separase‐WT and with OHT to induce nuclear accumulation of ER‐AsiSI and infliction of DSBs were synchronized in G1‐ or G2‐phase and analyzed by IP–Western blotting (D) and by IFM for γH2AX‐ and Myc‐separase‐positive foci (see Fig EV2E). The quantification of the IFM in (E) shows averages (bars) of three independent experiments (dots) counting ≥ 100 cells each.
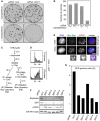
- A, B
Depletion of separase renders human cells hypersensitive to camptothecin. Twelve hours after transfection of the indicated siRNAs, 100 HEK293 cells each were plated onto 10‐cm petri dishes. Another 12 h later, camptothecin (0.25 nM end concentration) or carrier solvent (DMSO) was added. Colonies were stained by crystal violet 12 days thereafter and photographed (A). For each condition, three independent experiments were quantified by ImageJ (dots) and averaged (bars) (B).
- C
Schematic of the experiment shown in Figs 1D and E, and EV2D and E.
- D
Dox‐ and OHT‐treated HEK293 cells synchronized in G1‐ or G2‐phase were propidium iodide (PI)‐stained and analyzed by DNA content by flow cytometry.
- E
Representative IFM images of Dox‐ and OHT‐treated HEK293 cells synchronized in G1‐ or G2‐phase. Scale bar = 5 μm.
- F, G
U2OS DR‐GFP (HDR reporter) cells were transfected with GL2 or one of five different SEPARASE‐directed siRNAs. Following a second transfection to express ER‐tagged I‐SceI and addition of OHT to induce nuclear accumulation of the homing endonuclease, cells were subjected to flow cytometry (G) and immunoblotting (F).
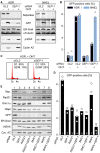
- A–C
Separase is required for proper HDR but dispensable for NHEJ. U2OS DR‐GFP (HDR reporter) and U2OS EJ5‐GFP (NHEJ reporter) cells were separase‐depleted by RNAi (SEP‐1) or control‐treated with GL2 siRNA, transfected to express HA‐tagged ER‐I‐SceI, and then supplemented with OHT in G2‐phase to induce nuclear accumulation of the homing endonuclease. Ethanol‐supplemented samples served as negative controls. Two days later, cells were subjected to immunoblotting (A) and flow cytometry to quantify the percentage of GFP‐positive cells (B) and PI‐stained cellular DNA (C). The GFP quantification in (B) displays averages (bars) of three independent experiments (dots).
- D, E
Co‐depletion of Wapl and separase has a synergistic effect on HDR. U2OS DR‐GFP cells were transfected with the indicated siRNAs, treated as in (A + B) and analyzed by GFP flow cytometry (D) and immunoblotting (E). Shown in (D) are averages (bars) of two to three independent experiments (dots).

Transgenic HEK293 cells constitutively expressing a separase sensor (cartoon below) and inducibly expressing Myc‐separase were transiently transfected to express Flag‐AsiSI‐ER, Dox‐ and/or OHT‐treated in G2‐phase, and analyzed by IP–Western blotting.
Cells from (A) were analyzed by IP–Western blotting for cleavage of endogenous Rad21. As a control, in vitro‐translated (IVT) Rad21 was incubated with hyperactive separase‐SA or a protease‐dead (PD) variant (Boos et al, 2008).
Sensor‐expressing cells were treated in G2‐phase with DRB and nocodazole for 6 and 2 h, respectively, prior to chromosome spreading and IFM using Hec1 and γH2AX antibodies. The separase sensor was detected based on autofluorescence of eGFP and mCherry, while Hec1 and γH2AX antibodies were detected with corresponding Cy5‐ and marina blue‐labeled secondary antibodies, respectively. Note that sizes of spread chromosomes vary greatly with buffer conditions, which is why no scale bar is shown.

- A–C
Ser1660 is phosphorylated in response to DNA damage and required for the interaction of separase with γH2AX. HEK293 cells were arrested in G2‐phase by sequential thymidine and RO‐3306 treatment, DRB‐ (+) or mock‐treated (−), and then analyzed as indicated. (A and B) Myc‐separase‐WT or Myc‐separase‐S1660A‐expressing cells were subjected to IP–Western blotting using, among others, a pan‐specific antibody against phosphorylated serine (A, pan‐pS) or a separase antibody specific for phosphorylated Ser1660 (B, pS1660). (C) DNA damage‐induced Ser1660 phosphorylation of endogenous separase is largely blocked by ATM inhibition. G2‐enriched HEK293 cells were treated with KU‐55933 (0.3 μM) and/or DRB and analyzed by IP–Western blotting 12 h thereafter using the indicated antibodies.
- D
Preventing NES phosphorylation spoils nuclear localization of separase in response to DSBs. HeLaK cells expressing N‐terminally NLS‐eGFP‐tagged separase variants were treated with DRB or carrier solvent (− DRB) for 4 h and then subjected to IFM using anti‐γH2AX and anti‐Nup153 to visualize sites of DNA damage and nuclear pore complexes, respectively. Transgenic separase was detected based on the eGFP autofluorescence. Note that due to their relatively high nuclear concentration, co‐localization of separase‐WT and separase‐S1660D with γH2AX foci is not discernable. Scale bar = 5 μm.
- E
In vitro phosphorylation of separase on Ser1660 by ATM kinase. Incubation of GST‐p53 (amino acids 9–22), GST, separase‐WT, or separase‐S1660A with recombinant ATM‐WT, ATM‐KD (kinase dead), and/or KU‐55933 in the presence of [γ‐33P]‐ATP was followed by immunoblotting and autoradiography.
- F
Arg‐methylation of RG‐repeats mediates recruitment of separase to DSB‐containing chromatin. Myc‐separase‐WT‐ or Myc‐separase‐KG‐expressing cells were treated with DRB as indicated and analyzed by IP–Western blotting and Coomassie staining.
- G
In vitro Arg‐methylation of separase's RG‐repeats by PRMT1. Incubation of histone H4, separase‐WT, or separase‐KG with recombinant PRMT1 or reference buffer in the presence of S‐adenosyl‐L‐[methyl‐3H]‐methionine was followed by Coomassie staining and autoradiography.

- A, B
HeLaK cells expressing N‐terminally NLS‐eGFP‐tagged separase‐WT or separase‐S1160D were treated with DRB, KU‐55933 (2 μM), or carrier solvent (−) for 4 h as indicated and then subjected to IFM and Western blot analysis using the indicated antibodies. Transgenic separase was detected based on the eGFP autofluorescence. Note that due to their relatively high nuclear concentration, co‐localization of separase‐WT and separase‐S1660D with γH2AX foci is not discernable. Scale bar = 5 μm.
- C, D
ATM enhances separase‐dependent Rad21 cleavage in vitro. Rad21 was 35S‐labeled by in vitro expression in reticulocyte lysate and treated with recombinant ATM kinase in the presence of KU‐55933 (10 μM) and/or ATP. Then, samples were incubated with recombinant separase and finally analyzed by SDS–PAGE and autoradiography. Wild‐type (WT) or kinase‐dead (KD) variants of ATM and hyperactive (SA) or protease‐dead (PD) variants of separase were employed as indicated.

Persistence of γH2AX‐ and MDC1‐positive foci in the absence of NES phosphorylation or RG‐methylation of separase. Transfected HEK293T cells were siRNA‐ and Dox‐treated to deplete endogenous separase and induce expression of the indicated, siRNA‐resistant Myc‐tagged separase variants, respectively. Then, they were constitutively (− washout) or transiently exposed to DRB (+ washout) and finally quantitatively assessed by IFM for γH2AX‐ and MDC1‐positive foci. Shown are averages (bars) of three independent experiments (dots) counting ≥ 100 cells each. Scale bar = 5 μm.
Schematic of the experiment.
Recruitment of separase‐WT and separase‐S1660D but not separase‐superNES, separase‐S1660A, and separase‐KG to DSBs. Cells from (A) were treated as indicated and analyzed by IFM for co‐localization of Myc‐separase with γH2AX foci. Scale bar = 5 μm.
Interaction of separase‐WT and separase‐S1660D but not separase‐superNES, separase‐S1660A, and separase‐KG with γH2AX. Cells from (A) were subjected to IP–Western blot analysis as indicated. Lanes that illustrate cellular levels of cyclin A2 and γH2AX after DRB washout and those that analyze interaction of Myc‐separase variants with γH2AX in the presence of DRB are labeled by red arrows and blue arrowheads, respectively.

- A, B
Separase is sumoylated in response to DSBs. HEK293T cells transfected to express His6‐separase (A) or His6‐Sumo2 (B) were treated with DRB or carrier solvent (− DRB) and then subjected to denaturing IMAC followed by immunoblotting of input samples and eluates using the indicated antibodies.
- C, D
Lys‐1034 is a major target of DSB‐induced sumoylation of separase. HEK293T cells expressing His6‐Sumo2 (C) or Myc‐separase‐WT or separase‐K1034R (D) were DRB‐ or mock‐treated and subjected to denaturing IMAC or Myc‐IP, respectively. Input samples and eluates were immunoblotted using the indicated antibodies.
- E
Mms21‐dependent in vitro sumoylation of separase at Lys‐1034. Myc beads were loaded with separase‐WT or separase‐K1034R or left empty (last lane), combined with recombinant Ubc9 and/or Mms21, as indicated, and incubated in the presence of His6‐Sumo2, Sae1‐Sae2, and ATP. Supernatant and washed beads were analyzed by Coomassie staining (lower panels) and immunoblotting (upper panels), respectively.
- F
Cells expressing the indicated separase variants were DRB‐ or mock‐treated and then lysed. Lysates and chromatin pelleted therefrom were assessed by immunoblotting and Coomassie staining, as indicated. Note that the lanes shown in the upper panels, although not directly juxtaposed, nevertheless originate from the same gel.
- G
Preventing sumoylation of Lys‐1034 compromises separase's ability to support DSB repair. The indicated variants were probed for their ability to functionally replace endogenous separase in HDR as described in Fig 6A and B. Shown are averages (bars) of three independent experiments (dots) counting ≥ 100 cells each.
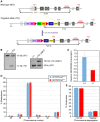
Diagrammatic representation of the relevant 5′ region of the SEPARASE locus. Exons are depicted as boxes with coding regions in gray. As indicated, the start (ATG) and stop (TGA) codons are located within exons E2 and E31, respectively. The regulating cassette from the TriTAUBi‐Bd plasmid is enclosed by a rtTS‐tTA cassette (Hayakawa et al, 2006). The selection markers NEO and URA are flanked by loxP sites (indicated by a P within red arrowheads). The minimum cytomegalovirus promoter (CMV), the tetO repeated sequences (tetO), all EcoRV (RV) sites, and the position of the Southern probe (see B) are indicated.
Southern blot analysis of EcoRV‐digested genomic DNA from tails of wild‐type (WT = +) mice and those with the targeted mutation (TG) before and after Cre‐mediated deletion of the selection cassette (TG ΔNEO). See (A) for expected sizes of EcoRV fragments.
Separase expression is halved in SEPARASE +/− MEFs. Northern blot analysis and densitometry were used to quantify the expression of SEPARASE mRNA normalized to the expression of ACTIN in total RNA preparations from SEPARASE +/+ versus SEPARASE +/− MEFs (n = 4). Values represent means ± standard deviations.
No increase in aneuploidy in SEPARASE +/− MEFs. MEFs were treated with colcemid for 4 h and processed using standard methanol–acetic fixation followed by DAPI staining. MEFs from three embryos of each genotype and more than 100 metaphases per sample were counted. Values represent means ± standard deviations.
No increase in lagging chromosomes in SEPARASE +/− MEFs. Cells passaged the day before were directly fixed and stained with DAPI. MEFs from three embryos of each genotype and more than 100 anaphases per sample were counted. Values represent means ± standard deviations.
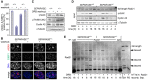
- A, B
SEPARASE +/+ and SEPARASE +/− MEFs were constitutively (− washout) or transiently exposed to DRB (+ washout) and then analyzed by IF for cells with γH2AX‐ and 53BP1‐positive foci. Shown are averages (bars) of three independent experiments (dots) counting ≥ 100 cells each. Scale bar = 5 μm.
- C
Cell lysates from one experiment in (A) were analyzed by immunoblotting using the indicated antibodies.
- D, E
SEPARASE +/+ and SEPARASE +/− MEFs were treated with DRB (+) or carrier solvent (−) for the indicated times and then analyzed by immunoblotting of total cell lysates (D) and IPs using anti‐Rad21 or unspecific IgG (E). The lower right two lanes show in vitro‐expressed mouse Rad21 treated with hyperactive (SA) separase or a protease‐dead (PD) variant.

Transformation susceptibility of primary MEFs. SEPARASE +/+ versus SEPARASE +/− MEFs were infected to express E1A and hRasV12 and then subjected to a colony formation assay. Shown are representative images (left) and the graphical representation of the number of transformed foci (right) from two independent experiments (bars = standard deviations).
Two‐stage DMBA (initiation) plus TPA (promotion) skin carcinogenesis. The number and size of skin papillomas are plotted against time for each genotype: SEPARASE +/+ (n = 8) versus SEPARASE +/− (n = 11).
Representative hematoxylin‐ and eosin‐stained sections from SEPARASE +/− biopsies. (i) Glandular hyperplasia; (ii) benign papilloma; (iii) squamous cell carcinoma. Scale bars are 200 μm in (i) and 500 μm in (ii) and (iii).
Quantitative histological assessment of the DMBA‐TPA skin carcinogenesis assay. Given are numbers of total, and corresponding percentage in brackets.
Similar articles
-
TIS21(/BTG2/PC3) accelerates the repair of DNA double strand breaks by enhancing Mre11 methylation and blocking damage signal transfer to the Chk2(T68)-p53(S20) pathway.DNA Repair (Amst). 2012 Dec 1;11(12):965-75. doi: 10.1016/j.dnarep.2012.09.009. Epub 2012 Oct 22. DNA Repair (Amst). 2012. PMID: 23089312
-
The influence of heterochromatin on DNA double strand break repair: Getting the strong, silent type to relax.DNA Repair (Amst). 2010 Dec 10;9(12):1273-82. doi: 10.1016/j.dnarep.2010.09.013. Epub 2010 Oct 30. DNA Repair (Amst). 2010. PMID: 21036673 Review.
-
The complexity of DNA double strand break is a crucial factor for activating ATR signaling pathway for G2/M checkpoint regulation regardless of ATM function.DNA Repair (Amst). 2015 Jan;25:72-83. doi: 10.1016/j.dnarep.2014.11.004. Epub 2014 Nov 24. DNA Repair (Amst). 2015. PMID: 25497328
-
ATM Paradoxically Promotes Oncogenic Transformation via Transcriptional Reprogramming.Cancer Res. 2020 Apr 15;80(8):1669-1680. doi: 10.1158/0008-5472.CAN-19-2255. Epub 2020 Feb 14. Cancer Res. 2020. PMID: 32060145 Free PMC article.
-
Double strand break (DSB) repair in heterochromatin and heterochromatin proteins in DSB repair.DNA Repair (Amst). 2014 Jul;19:163-8. doi: 10.1016/j.dnarep.2014.03.015. Epub 2014 Apr 19. DNA Repair (Amst). 2014. PMID: 24754998 Review.
Cited by
-
Mapping separase-mediated cleavage in situ.NAR Genom Bioinform. 2022 Nov 18;4(4):lqac085. doi: 10.1093/nargab/lqac085. eCollection 2022 Dec. NAR Genom Bioinform. 2022. PMID: 36415827 Free PMC article.
-
PRMT1 Modulates Alternative Splicing to Enhance HPV18 mRNA Stability and Promote the Establishment of Infection.bioRxiv [Preprint]. 2024 Sep 26:2024.09.26.614592. doi: 10.1101/2024.09.26.614592. bioRxiv. 2024. PMID: 39386465 Free PMC article. Preprint.
-
Eliminating separase inhibition reveals absence of robust cohesin protection in oocyte metaphase II.EMBO J. 2025 Aug 5. doi: 10.1038/s44318-025-00522-0. Online ahead of print. EMBO J. 2025. PMID: 40764838
-
Live Cell Monitoring of Separase Activity, a Key Enzymatic Reaction for Chromosome Segregation, with Chimeric FRET-Based Molecular Sensor upon Cell Cycle Progression.Biosensors (Basel). 2024 Apr 15;14(4):192. doi: 10.3390/bios14040192. Biosensors (Basel). 2024. PMID: 38667185 Free PMC article.
-
Separase activity distribution can be a marker of major molecular response and proliferation of CD34+ cells in TKI-treated chronic myeloid leukemia patients.Ann Hematol. 2020 May;99(5):991-1006. doi: 10.1007/s00277-020-04007-4. Epub 2020 Apr 6. Ann Hematol. 2020. PMID: 32253454 Free PMC article. Clinical Trial.
References
-
- Boos D, Kuffer C, Lenobel R, Korner R, Stemmann O (2008) Phosphorylation‐dependent binding of cyclin B1 to a Cdc6‐like domain of human separase. J Biol Chem 283: 816–823 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

