The Genetic Transformation of Chlamydia pneumoniae
- PMID: 30305318
- PMCID: PMC6180227
- DOI: 10.1128/mSphere.00412-18
The Genetic Transformation of Chlamydia pneumoniae
Abstract
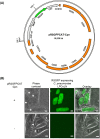
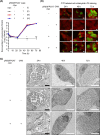
We demonstrate the genetic transformation of Chlamydia pneumoniae using a plasmid shuttle vector system which generates stable transformants. The equine C. pneumoniae N16 isolate harbors the 7.5-kb plasmid pCpnE1. We constructed the plasmid vector pRSGFPCAT-Cpn containing a pCpnE1 backbone, plus the red-shifted green fluorescent protein (RSGFP), as well as the chloramphenicol acetyltransferase (CAT) gene used for the selection of plasmid shuttle vector-bearing C. pneumoniae transformants. Using the pRSGFPCAT-Cpn plasmid construct, expression of RSGFP in koala isolate C. pneumoniae LPCoLN was demonstrated. Furthermore, we discovered that the human cardiovascular isolate C. pneumoniae CV-6 and the human community-acquired pneumonia-associated C. pneumoniae IOL-207 could also be transformed with pRSGFPCAT-Cpn. In previous studies, it was shown that Chlamydia spp. cannot be transformed when the plasmid shuttle vector is constructed from a different plasmid backbone to the homologous species. Accordingly, we confirmed that pRSGFPCAT-Cpn could not cross the species barrier in plasmid-bearing and plasmid-free C. trachomatis, C. muridarum, C. caviae, C. pecorum, and C. abortus However, contrary to our expectation, pRSGFPCAT-Cpn did transform C. felis Furthermore, pRSGFPCAT-Cpn did not recombine with the wild-type plasmid of C. felis Taken together, we provide for the first time an easy-to-handle transformation protocol for C. pneumoniae that results in stable transformants. In addition, the vector can cross the species barrier to C. felis, indicating the potential of horizontal pathogenic gene transfer via a plasmid.IMPORTANCE The absence of tools for the genetic manipulation of C. pneumoniae has hampered research into all aspects of its biology. In this study, we established a novel reproducible method for C. pneumoniae transformation based on a plasmid shuttle vector system. We constructed a C. pneumoniae plasmid backbone shuttle vector, pRSGFPCAT-Cpn. The construct expresses the red-shifted green fluorescent protein (RSGFP) fused to chloramphenicol acetyltransferase in C. pneumoniaeC. pneumoniae transformants stably retained pRSGFPCAT-Cpn and expressed RSGFP in epithelial cells, even in the absence of chloramphenicol. The successful transformation in C. pneumoniae using pRSGFPCAT-Cpn will advance the field of chlamydial genetics and is a promising new approach to investigate gene functions in C. pneumoniae biology. In addition, we demonstrated that pRSGFPCAT-Cpn overcame the plasmid species barrier without the need for recombination with an endogenous plasmid, indicating the potential probability of horizontal chlamydial pathogenic gene transfer by plasmids between chlamydial species.
Keywords: Chlamydia felis; Chlamydia pneumoniae; genetic manipulation; plasmid shuttle vector; plasmid tropism; transformation.
Copyright © 2018 Shima et al.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
