A 1,000-Year-Old RNA Virus
- PMID: 30305356
- PMCID: PMC6288340
- DOI: 10.1128/JVI.01188-18
A 1,000-Year-Old RNA Virus
Abstract
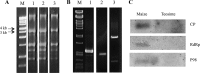
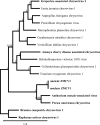
Only a few RNA viruses have been discovered from archaeological samples, the oldest dating from about 750 years ago. Using ancient maize cobs from Antelope house, Arizona, dating from ca. 1,000 CE, we discovered a novel plant virus with a double-stranded RNA genome. The virus is a member of the family Chrysoviridae that infect plants and fungi in a persistent manner. The extracted double-stranded RNA from 312 maize cobs was converted to cDNA, and sequences were determined using an Illumina HiSeq 2000. Assembled contigs from many samples showed similarity to Anthurium mosaic-associated virus and Persea americana chrysovirus, putative species in the Chrysovirus genus, and nearly complete genomes were found in three ancient maize samples. We named this new virus Zea mays chrysovirus 1. Using specific primers, we were able to recover sequences of a closely related virus from modern maize and obtained the nearly complete sequences of the three genomic RNAs. Comparing the nucleotide sequences of the three genomic RNAs of the modern and ancient viruses showed 98, 96.7, and 97.4% identities, respectively. Hence, in 1,000 years of maize cultivation, this virus has undergone about 3% divergence.IMPORTANCE A virus related to plant chrysoviruses was found in numerous ancient samples of maize, with nearly complete genomes in three samples. The age of the ancient samples (i.e., about 1,000 years old) was confirmed by carbon dating. Chrysoviruses are persistent plant viruses. They infect their hosts from generation to generation by transmission through seeds and can remain in their hosts for very long time periods. When modern corn samples were analyzed, a closely related chrysovirus was found with only about 3% divergence from the ancient sequences. This virus represents the oldest known plant virus.
Keywords: ancient tissue; maize; virus evolution.
Copyright © 2018 American Society for Microbiology.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

