LRH-1 mitigates intestinal inflammatory disease by maintaining epithelial homeostasis and cell survival
- PMID: 30305617
- PMCID: PMC6180039
- DOI: 10.1038/s41467-018-06137-w
LRH-1 mitigates intestinal inflammatory disease by maintaining epithelial homeostasis and cell survival
Abstract
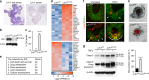
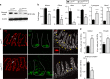
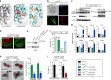
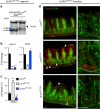
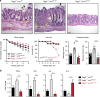
Epithelial dysfunction and crypt destruction are defining features of inflammatory bowel disease (IBD). However, current IBD therapies targeting epithelial dysfunction are lacking. The nuclear receptor LRH-1 (NR5A2) is expressed in intestinal epithelium and thought to contribute to epithelial renewal. Here we show that LRH-1 maintains intestinal epithelial health and protects against inflammatory damage. Knocking out LRH-1 in murine intestinal organoids reduces Notch signaling, increases crypt cell death, distorts the cellular composition of the epithelium, and weakens the epithelial barrier. Human LRH-1 (hLRH-1) rescues epithelial integrity and when overexpressed, mitigates inflammatory damage in murine and human intestinal organoids, including those derived from IBD patients. Finally, hLRH-1 greatly reduces disease severity in T-cell-mediated murine colitis. Together with the failure of a ligand-incompetent hLRH-1 mutant to protect against TNFα-damage, these findings provide compelling evidence that hLRH-1 mediates epithelial homeostasis and is an attractive target for intestinal disease.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

