Genome-wide association studies of brain imaging phenotypes in UK Biobank
- PMID: 30305740
- PMCID: PMC6786974
- DOI: 10.1038/s41586-018-0571-7
Genome-wide association studies of brain imaging phenotypes in UK Biobank
Abstract
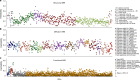
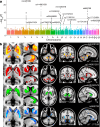
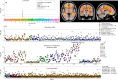
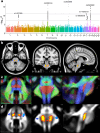
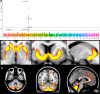
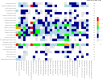
The genetic architecture of brain structure and function is largely unknown. To investigate this, we carried out genome-wide association studies of 3,144 functional and structural brain imaging phenotypes from UK Biobank (discovery dataset 8,428 subjects). Here we show that many of these phenotypes are heritable. We identify 148 clusters of associations between single nucleotide polymorphisms and imaging phenotypes that replicate at P < 0.05, when we would expect 21 to replicate by chance. Notable significant, interpretable associations include: iron transport and storage genes, related to magnetic susceptibility of subcortical brain tissue; extracellular matrix and epidermal growth factor genes, associated with white matter micro-structure and lesions; genes that regulate mid-line axon development, associated with organization of the pontine crossing tract; and overall 17 genes involved in development, pathway signalling and plasticity. Our results provide insights into the genetic architecture of the brain that are relevant to neurological and psychiatric disorders, brain development and ageing.
Conflict of interest statement
J.M. is a co-founder and director of GENSCI Ltd. S.M.S. is a co-founder of SBGneuro. The other authors declare no competing interests.
Figures
Comment in
-
UK Biobank shares the promise of big data.Nature. 2018 Oct;562(7726):194-195. doi: 10.1038/d41586-018-06948-3. Nature. 2018. PMID: 30305754 No abstract available.
-
UK Biobank data on 500,000 people paves way to precision medicine.Nature. 2018 Oct;562(7726):163-164. doi: 10.1038/d41586-018-06950-9. Nature. 2018. PMID: 30365879 No abstract available.
References
-
- Toga, A. W. Brain Mapping: An Encyclopedic Reference (Academic, Amsterdam, 2015).
-
- Allen, N. et al. UK Biobank: Current status and what it means for epidemiology. Health Policy Technol. 1, 123–126 (2012). - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

