Multigene panel testing beyond BRCA1/2 in breast/ovarian cancer Spanish families and clinical actionability of findings
- PMID: 30306255
- PMCID: PMC11813382
- DOI: 10.1007/s00432-018-2763-9
Multigene panel testing beyond BRCA1/2 in breast/ovarian cancer Spanish families and clinical actionability of findings
Abstract
Purpose: Few and small studies have been reported about multigene testing usage by massively parallel sequencing in European cancer families. There is an open debate about what genes should be tested, and the actionability of some included genes is under research.
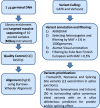
Methods: We investigated a panel of 34 known high/moderate-risk cancer genes, including 16 related to breast or ovarian cancer (BC/OC) genes, and 63 candidate genes to BC/OC in 192 clinically suspicious of hereditary breast/ovarian cancer (HBOC) Spanish families without pathogenic variants in BRCA1 or BRCA2 (BRCA1/2).
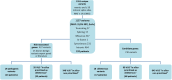
Results: We identified 16 patients who carried a high- or moderate-risk pathogenic variant in eight genes: 4 PALB2, 3 ATM, 2 RAD51D, 2 TP53, 2 APC, 1 BRIP1, 1 PTEN and 1 PMS2. These findings led to increased surveillance or prevention options in 12 patients and predictive testing in their family members. We detected 383 unique variants of uncertain significance in known cancer genes, of which 35 were prioritized in silico. Eighteen loss-of-function variants were detected in candidate BC/OC genes in 17 patients (1 BARD1, 1 ERCC3, 1 ERCC5, 2 FANCE, 1 FANCI, 2 FANCL, 1 FANCM, 1 MCPH1, 1 PPM1D, 2 RBBP8, 3 RECQL4 and 1 with SLX4 and XRCC2), three of which also carry pathogenic variants in known cancer genes.
Conclusions: Eight percent of the BRCA1/2 negative patients carry pathogenic variants in other actionable genes. The multigene panel usage improves the diagnostic yield in HBOC testing and it is an effective tool to identify potentially new candidate genes.
Keywords: BRCA1 or BRCA2 negative; Breast/ovarian hereditary cancer; Clinical actionability; Panel testing by massively parallel sequencing.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Balmaña J, Balaguer F, Cervantes A, Arnold D (2013) Familial risk-colorectal cancer: ESMO clinical practice guidelines. Ann Oncol 24:vi73–vi80. 10.1093/annonc/mdt209 - PubMed
-
- Borràs E, Pineda M, Cadiñanos J et al (2013) Refining the role of pms2 in Lynch syndrome: germline mutational analysis improved by comprehensive assessment of variants. J Med Genet 50:552–563. 10.1136/jmedgenet-2012-101511 - PubMed
-
- Bouaoun L, Sonkin D, Ardin M et al (2016) TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum Mutat 37:865–876. 10.1002/humu.23035 - PubMed
-
- Buys SS, Sandbach JF, Gammon A et al (2017) A study of over 35,000 women with breast cancer tested with a 25-gene panel of hereditary cancer genes. Cancer 123:1721–1730. 10.1002/cncr.30498 - PubMed
-
- Cardoso M, Paulo P, Maia S, Teixeira MR (2016) Truncating and missense PPM1D mutations in early-onset and/or familial/hereditary prostate cancer patients. Genes Chromosom Cancer 55:954–961. 10.1002/gcc.22393 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

