Explicit ions/implicit water generalized Born model for nucleic acids
- PMID: 30307229
- PMCID: PMC5959738
- DOI: 10.1063/1.5027260
Explicit ions/implicit water generalized Born model for nucleic acids
Abstract
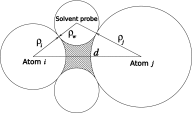
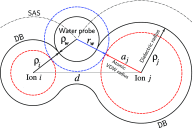
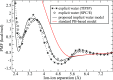
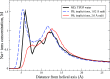
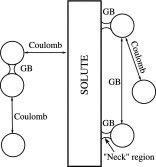
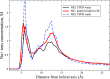
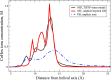
The ion atmosphere around highly charged nucleic acid molecules plays a significant role in their dynamics, structure, and interactions. Here we utilized the implicit solvent framework to develop a model for the explicit treatment of ions interacting with nucleic acid molecules. The proposed explicit ions/implicit water model is based on a significantly modified generalized Born (GB) model and utilizes a non-standard approach to define the solute/solvent dielectric boundary. Specifically, the model includes modifications to the GB interaction terms for the case of multiple interacting solutes-disconnected dielectric boundary around the solute-ion or ion-ion pairs. A fully analytical description of all energy components for charge-charge interactions is provided. The effectiveness of the approach is demonstrated by calculating the potential of mean force for Na+-Cl- ion pair and by carrying out a set of Monte Carlo (MC) simulations of mono- and trivalent ions interacting with DNA and RNA duplexes. The monovalent (Na+) and trivalent (CoHex3+) counterion distributions predicted by the model are in close quantitative agreement with all-atom explicit water molecular dynamics simulations used as reference. Expressed in the units of energy, the maximum deviations of local ion concentrations from the reference are within k B T. The proposed explicit ions/implicit water GB model is able to resolve subtle features and differences of CoHex distributions around DNA and RNA duplexes. These features include preferential CoHex binding inside the major groove of the RNA duplex, in contrast to CoHex biding at the "external" surface of the sugar-phosphate backbone of the DNA duplex; these differences in the counterion binding patters were earlier shown to be responsible for the observed drastic differences in condensation propensities between short DNA and RNA duplexes. MC simulations of CoHex ions interacting with the homopolymeric poly(dA·dT) DNA duplex with modified (de-methylated) and native thymine bases are used to explore the physics behind CoHex-thymine interactions. The simulations suggest that the ion desolvation penalty due to proximity to the low dielectric volume of the methyl group can contribute significantly to CoHex-thymine interactions. Compared to the steric repulsion between the ion and the methyl group, the desolvation penalty interaction has a longer range and may be important to consider in the context of methylation effects on DNA condensation.
Figures

Similar articles
-
Implicit Solvent with Explicit Ions Generalized Born Model in Molecular Dynamics: Application to DNA.J Chem Theory Comput. 2024 Oct 8;20(19):8724-8739. doi: 10.1021/acs.jctc.4c00833. Epub 2024 Sep 16. J Chem Theory Comput. 2024. PMID: 39283928 Free PMC article.
-
Generalized Born Implicit Solvent Models for Biomolecules.Annu Rev Biophys. 2019 May 6;48:275-296. doi: 10.1146/annurev-biophys-052118-115325. Epub 2019 Mar 11. Annu Rev Biophys. 2019. PMID: 30857399 Free PMC article. Review.
-
Multi-shell model of ion-induced nucleic acid condensation.J Chem Phys. 2016 Apr 21;144(15):155101. doi: 10.1063/1.4945382. J Chem Phys. 2016. PMID: 27389241 Free PMC article.
-
Explicit ion, implicit water solvation for molecular dynamics of nucleic acids and highly charged molecules.J Comput Chem. 2008 May;29(7):1113-30. doi: 10.1002/jcc.20874. J Comput Chem. 2008. PMID: 18074338
-
The ABCs of molecular dynamics simulations on B-DNA, circa 2012.J Biosci. 2012 Jul;37(3):379-97. doi: 10.1007/s12038-012-9222-6. J Biosci. 2012. PMID: 22750978 Free PMC article. Review.
Cited by
-
Implicit Solvent with Explicit Ions Generalized Born Model in Molecular Dynamics: Application to DNA.J Chem Theory Comput. 2024 Oct 8;20(19):8724-8739. doi: 10.1021/acs.jctc.4c00833. Epub 2024 Sep 16. J Chem Theory Comput. 2024. PMID: 39283928 Free PMC article.
-
Graph deep learning locates magnesium ions in RNA.QRB Discov. 2022;3:e20. doi: 10.1017/qrd.2022.17. Epub 2022 Oct 6. QRB Discov. 2022. PMID: 37292390 Free PMC article.
-
Generalized Born Implicit Solvent Models for Biomolecules.Annu Rev Biophys. 2019 May 6;48:275-296. doi: 10.1146/annurev-biophys-052118-115325. Epub 2019 Mar 11. Annu Rev Biophys. 2019. PMID: 30857399 Free PMC article. Review.
-
Uncovering water effects in protein-ligand recognition: importance in the second hydration shell and binding kinetics.Phys Chem Chem Phys. 2023 Jan 18;25(3):2098-2109. doi: 10.1039/d2cp04584b. Phys Chem Chem Phys. 2023. PMID: 36562309 Free PMC article.
-
Structure and Zeta Potential of Gold Nanoparticles with Coronas of Varying Size and Composition.J Phys Chem C Nanomater Interfaces. 2025 Feb 18;129(8):4204-4214. doi: 10.1021/acs.jpcc.4c07595. eCollection 2025 Feb 27. J Phys Chem C Nanomater Interfaces. 2025. PMID: 40041390
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

