A gene expression comparison of Trypanosoma brucei and Trypanosoma congolense in the bloodstream of the mammalian host reveals species-specific adaptations to density-dependent development
- PMID: 30307943
- PMCID: PMC6199001
- DOI: 10.1371/journal.pntd.0006863
A gene expression comparison of Trypanosoma brucei and Trypanosoma congolense in the bloodstream of the mammalian host reveals species-specific adaptations to density-dependent development
Abstract
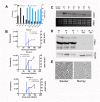
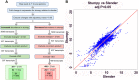
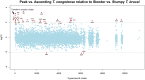
In the bloodstream of mammalian hosts Trypanosoma brucei undergoes well-characterised density-dependent growth control and developmental adaptation for transmission. This involves the differentiation from proliferative, morphologically 'slender' forms to quiescent 'stumpy' forms that preferentially infect the tsetse fly vector. Another important livestock trypanosome, Trypanosoma congolense, also undergoes density-dependent cell-cycle arrest although this is not linked to obvious morphological transformation. Here we have compared the gene expression profile of T. brucei and T. congolense during the ascending phase of the parasitaemia and at peak parasitaemia in mice, analysing species and developmental differences between proliferating and cell-cycle arrested forms. Despite underlying conservation of their quorum sensing signalling pathway, each species exhibits distinct profiles of gene regulation when analysed by orthogroup and cell surface phylome profiling. This analysis of peak parasitaemia T. congolense provides the first molecular signatures of potential developmental competence, assisting life cycle developmental studies in these important livestock parasites. Furthermore, comparison with T. brucei identifies candidate molecules from each species that may be important for their survival in the mammalian host, transmission or distinct tropism in the tsetse vector.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







Similar articles
-
Interspecies quorum sensing in co-infections can manipulate trypanosome transmission potential.Nat Microbiol. 2017 Nov;2(11):1471-1479. doi: 10.1038/s41564-017-0014-5. Epub 2017 Sep 4. Nat Microbiol. 2017. PMID: 28871083 Free PMC article.
-
Sequential infection of tsetse flies with Trypanosoma congolense and Trypanosoma brucei.Acta Trop. 1992 Apr;50(4):345-52. doi: 10.1016/0001-706x(92)90070-e. Acta Trop. 1992. PMID: 1356306
-
Genome-wide dissection of the quorum sensing signalling pathway in Trypanosoma brucei.Nature. 2014 Jan 30;505(7485):681-685. doi: 10.1038/nature12864. Epub 2013 Dec 15. Nature. 2014. PMID: 24336212 Free PMC article.
-
Trypanosoma congolense: Molecular Toolkit and Resources for Studying a Major Livestock Pathogen and Model Trypanosome.Adv Parasitol. 2017;98:283-309. doi: 10.1016/bs.apar.2017.03.002. Epub 2017 May 5. Adv Parasitol. 2017. PMID: 28942771 Review.
-
Bloodstream form pre-adaptation to the tsetse fly in Trypanosoma brucei.Front Cell Infect Microbiol. 2013 Nov 14;3:78. doi: 10.3389/fcimb.2013.00078. eCollection 2013. Front Cell Infect Microbiol. 2013. PMID: 24294594 Free PMC article. Review.
Cited by
-
A conserved trypanosomatid differentiation regulator controls substrate attachment and morphological development in Trypanosoma congolense.PLoS Pathog. 2024 Feb 26;20(2):e1011889. doi: 10.1371/journal.ppat.1011889. eCollection 2024 Feb. PLoS Pathog. 2024. PMID: 38408115 Free PMC article.
-
Trypanosoma brucei brucei Induces Polymorphonuclear Neutrophil Activation and Neutrophil Extracellular Traps Release.Front Immunol. 2020 Oct 22;11:559561. doi: 10.3389/fimmu.2020.559561. eCollection 2020. Front Immunol. 2020. PMID: 33193328 Free PMC article.
-
Developmental competence and antigen switch frequency can be uncoupled in Trypanosoma brucei.Proc Natl Acad Sci U S A. 2019 Nov 5;116(45):22774-22782. doi: 10.1073/pnas.1912711116. Epub 2019 Oct 21. Proc Natl Acad Sci U S A. 2019. PMID: 31636179 Free PMC article.
-
A potential role for a novel ZC3H5 complex in regulating mRNA translation in Trypanosoma brucei.J Biol Chem. 2020 Oct 16;295(42):14291-14304. doi: 10.1074/jbc.RA120.014346. Epub 2020 Aug 5. J Biol Chem. 2020. PMID: 32763974 Free PMC article.
-
Several different sequences are implicated in bloodstream-form-specific gene expression in Trypanosoma brucei.PLoS Negl Trop Dis. 2022 Mar 21;16(3):e0010030. doi: 10.1371/journal.pntd.0010030. eCollection 2022 Mar. PLoS Negl Trop Dis. 2022. PMID: 35312693 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

