A polymorphic residue that attenuates the antiviral potential of interferon lambda 4 in hominid lineages
- PMID: 30308076
- PMCID: PMC6181419
- DOI: 10.1371/journal.ppat.1007307
A polymorphic residue that attenuates the antiviral potential of interferon lambda 4 in hominid lineages
Abstract
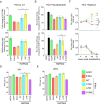
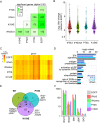
As antimicrobial signalling molecules, type III or lambda interferons (IFNλs) are critical for defence against infection by diverse pathogens, including bacteria, fungi and viruses. Counter-intuitively, expression of one member of the family, IFNλ4, is associated with decreased clearance of hepatitis C virus (HCV) in the human population; by contrast, a natural frameshift mutation that abrogates IFNλ4 production improves HCV clearance. To further understand how genetic variation between and within species affects IFNλ4 function, we screened a panel of all known extant coding variants of human IFNλ4 for their antiviral potential and identify three that substantially affect activity: P70S, L79F and K154E. The most notable variant was K154E, which was found in African Congo rainforest 'Pygmy' hunter-gatherers. K154E greatly enhanced in vitro activity in a range of antiviral (HCV, Zika virus, influenza virus and encephalomyocarditis virus) and gene expression assays. Remarkably, E154 is the ancestral residue in mammalian IFNλ4s and is extremely well conserved, yet K154 has been fixed throughout evolution of the hominid genus Homo, including Neanderthals. Compared to chimpanzee IFNλ4, the human orthologue had reduced activity due to amino acid K154. Comparison of published gene expression data from humans and chimpanzees showed that this difference in activity between K154 and E154 in IFNλ4 correlates with differences in antiviral gene expression in vivo during HCV infection. Mechanistically, our data show that the human-specific K154 negatively affects IFNλ4 activity through a novel means by reducing its secretion and potency. We thus demonstrate that attenuated activity of IFNλ4 is conserved among humans and postulate that differences in IFNλ4 activity between species contribute to distinct host-specific responses to-and outcomes of-infection, such as HCV infection. The driver of reduced IFNλ4 antiviral activity in humans remains unknown but likely arose between 6 million and 360,000 years ago in Africa.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008. 89(1):1–47. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

