Enlisting commensal microbes to resist antibiotic-resistant pathogens
- PMID: 30309968
- PMCID: PMC6314519
- DOI: 10.1084/jem.20180399
Enlisting commensal microbes to resist antibiotic-resistant pathogens
Abstract
The emergence of antibiotic-resistant bacterial pathogens is an all-too-common consequence of antibiotic use. Although antibiotic resistance among virulent bacterial pathogens is a growing concern, the highest levels of antibiotic resistance occur among less pathogenic but more common bacteria that are prevalent in healthcare settings. Patient-to-patient transmission of these antibiotic-resistant bacteria is a perpetual concern in hospitals. Many of these resistant microbes, such as vancomycin-resistant Enterococcus faecium and carbapenem-resistant Klebsiella pneumoniae, emerge from the intestinal lumen and invade the bloodstream of vulnerable patients, causing disseminated infection. These infections are associated with preceding antibiotic administration, which changes the intestinal microbiota and compromises resistance to colonization by antibiotic-resistant bacteria. Recent and ongoing studies are increasingly defining commensal bacterial species and the inhibitory mechanisms they use to prevent infection. The use of next-generation probiotics derived from the intestinal microbiota represents an alternative approach to prevention of infection by enriching colonization with protective commensal species, thereby reducing the density of antibiotic-resistant bacteria and also reducing patient-to-patient transmission of infection in healthcare settings.
© 2018 Keith and Pamer.
Figures
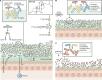
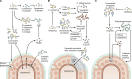
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

