ClinGen's GenomeConnect registry enables patient-centered data sharing
- PMID: 30311371
- PMCID: PMC6188701
- DOI: 10.1002/humu.23633
ClinGen's GenomeConnect registry enables patient-centered data sharing
Abstract
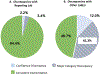
GenomeConnect, the NIH-funded Clinical Genome Resource (ClinGen) patient registry, engages patients in data sharing to support the goal of creating a genomic knowledge base to inform clinical care and research. Participant self-reported health information and genomic variants from genetic testing reports are curated and shared with public databases, such as ClinVar. There are four primary benefits of GenomeConnect: (1) sharing novel genomic data-47.9% of variants were new to ClinVar, highlighting patients as a genomic data source; (2) contributing additional phenotypic information-of the 52.1% of variants already in ClinVar, GenomeConnect provided enhanced case-level data; (3) providing a way for patients to receive variant classification updates if the reporting laboratory submits to ClinVar-97.3% of responding participants opted to receive such information and 13 updates have been identified; and (4) supporting connections with others, including other participants, clinicians, and researchers to enable the exchange of information and support-60.4% of participants have opted to partake in participant matching. Moving forward, ClinGen plans to increase patient-centric data sharing by partnering with other existing patient groups. By engaging patients, more information is contributed to the public knowledge base, benefiting both patients and the genomics community.
Keywords: ClinGen; ClinVar; genomic data sharing; matchmaking; patient registry; variant interpretation.
© 2018 Wiley Periodicals, Inc.
Figures



References
-
- American Medical Association. (2013). Genome Analysis and Variant Identification Policy D-460.971.
-
- American College of Medical Genetics (ACMG) Board of Directors. (2017). Laboratory and clinical genomic data sharing is crucial to improving genetic health care: a position statement of the American College of Medical Genetics and Genomics. Genetics in Medicine 19, 721–722. doi:10.1038/gim.2016.196 - DOI - PubMed
-
- Kirkpatrick BE, Riggs ER, Azzariti DR, Miller VR, Ledbetter DH, Miller DT, … Faucett WA on behalf of the ClinGen Resource. (2015) GenomeConnect: matchmaking between patients, clinical laboratories, and researchers to improve genomic knowledge. Human Mutation, 36, 974–978. doi: 10.1002/humu.22838 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

