ClinGen Allele Registry links information about genetic variants
- PMID: 30311374
- PMCID: PMC6519371
- DOI: 10.1002/humu.23637
ClinGen Allele Registry links information about genetic variants
Abstract
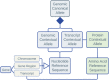
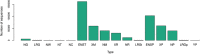
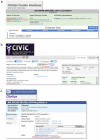
Effective exchange of information about genetic variants is currently hampered by the lack of readily available globally unique variant identifiers that would enable aggregation of information from different sources. The ClinGen Allele Registry addresses this problem by providing (1) globally unique "canonical" variant identifiers (CAids) on demand, either individually or in large batches; (2) access to variant-identifying information in a searchable Registry; (3) links to allele-related records in many commonly used databases; and (4) services for adding links to information about registered variants in external sources. A core element of the Registry is a canonicalization service, implemented using in-memory sequence alignment-based index, which groups variant identifiers denoting the same nucleotide variant and assigns unique and dereferenceable CAids. More than 650 million distinct variants are currently registered, including those from gnomAD, ExAC, dbSNP, and ClinVar, including a small number of variants registered by Registry users. The Registry is accessible both via a web interface and programmatically via well-documented Hypertext Transfer Protocol (HTTP) Representational State Transfer Application Programming Interface (REST-APIs). For programmatic interoperability, the Registry content is accessible in the JavaScript Object Notation for Linked Data (JSON-LD) format. We present several use cases and demonstrate how the linked information may provide raw material for reasoning about variant's pathogenicity.
Keywords: HGVS representation; linked data; pathogenicity of genetic variants; variant centric resources; variant identifiers.
© 2018 The Authors. Human Mutation published by Wiley Periodicals, Inc.
Conflict of interest statement
SEP is a member of the Baylor Genetics Scientific Advisory Panel. AM is an employee of BCM and performs integration consulting services for BCM‐developed software including Genboree through IP Genesis, Inc. LB is employed by Sunquest Information Systems company. Sunquest is a commercial laboratory software vendor. Other authors do not have any conflicts of interest. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Figures



References
-
- Fokkema, I. F. , Taschner, P. E. , Schaafsma, G. C. , Celli, J. , Laros, J. F. , & den Dunnen, J. T. (2011). LOVD v.2.0: The next generation in gene variant databases. Human Mutation, 32(5), 557–563. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

