The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark
- PMID: 30311405
- PMCID: PMC6492194
- DOI: 10.1002/wrna.1510
The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark
Abstract
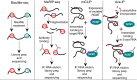
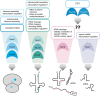
It is a well-known fact that RNA is the target of a plethora of modifications which currently amount to over a hundred. The vast majority of these modifications was observed in the two most abundant classes of RNA, rRNA and tRNA. With the recent advance in mapping technologies, modifications have been discovered also in mRNA and in less abundant non-coding RNA species. These developments have sparked renewed interest in elucidating the nature and functions of those "epitransciptomic" modifications in RNA. N6-methyladenosine (m6 A) is the best understood and most frequent mark of mRNA with demonstrated functions ranging from pre-mRNA processing, translation, miRNA biogenesis to mRNA decay. By contrast, much less research has been conducted on 5-methylcytosine (m5C), which was detected in tRNAs and rRNAs and more recently in poly(A)RNAs. In this review, we discuss recent developments in the discovery of m5C RNA methylomes, the functions of m5C as well as the proteins installing, translating and manipulating this modification. Although our knowledge about m5C in RNA transcripts is just beginning to consolidate, it has become clear that cytosine methylation represents a powerful mechanistic strategy to regulate cellular processes on an epitranscriptomic level. This article is categorized under: RNA Processing > RNA Editing and Modification RNA Interactions with Proteins and Other Molecules > Protein-RNA Interactions: Functional Implications RNA Processing > tRNA Processing RNA Turnover and Surveillance > Regulation of RNA Stability.
Keywords: 5-methylcytosine; N6-methyladenosine; RNA modification; epitranscriptomic mark; mRNA; miRNA; rRNA; tRNA.
© 2018 The Authors. WIREs RNA published by Wiley Periodicals, Inc.
Conflict of interest statement
The authors have declared no conflicts of interest for this article.
Figures

References
-
- Amos, H. , & Korn, M. (1958). 5‐Methyl cytosine in the RNA of Escherichia coli . Biochimica et Biophysica Acta, 29(2), 444–445. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

