Structural biology of G protein-coupled receptor signaling complexes
- PMID: 30311978
- PMCID: PMC6371222
- DOI: 10.1002/pro.3526
Structural biology of G protein-coupled receptor signaling complexes
Abstract
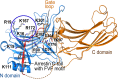
G protein-coupled receptors (GPCRs) constitute the largest family of cell surface receptors that mediate numerous cell signaling pathways, and are targets of more than one-third of clinical drugs. Thanks to the advancement of novel structural biology technologies, high-resolution structures of GPCRs in complex with their signaling transducers, including G-protein and arrestin, have been determined. These 3D complex structures have significantly improved our understanding of the molecular mechanism of GPCR signaling and provided a structural basis for signaling-biased drug discovery targeting GPCRs. Here we summarize structural studies of GPCR signaling complexes with G protein and arrestin using rhodopsin as a model system, and highlight the key features of GPCR conformational states in biased signaling including the sequence motifs of receptor TM6 that determine selective coupling of G proteins, and the phosphorylation codes of GPCRs for arrestin recruitment. We envision the future of GPCR structural biology not only to solve more high-resolution complex structures but also to show stepwise GPCR signaling complex assembly and disassembly and dynamic process of GPCR signal transduction.
Keywords: G protein; G protein-coupled receptors; GPCR; Structural biology; arrestin; signaling.
© 2018 The Protein Society.
Figures
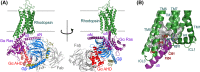




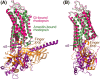
References
-
- Pierce KL, Premont RT, Lefkowitz RJ (2002) Seven‐transmembrane receptors. Nat Rev Mol Cell Biol 3:639–650. - PubMed
-
- Bjarnadottir TK, Gloriam DE, Hellstrand SH, Kristiansson H, Fredriksson R, Schioth HB (2006) Comprehensive repertoire and phylogenetic analysis of the G protein‐coupled receptors in human and mouse. Genomics 88:263–273. - PubMed
-
- Hamm HE (1998) The many faces of G protein signaling. J Biol Chem 273:669–672. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

