The somatic piRNA pathway controls germline transposition over generations
- PMID: 30312469
- PMCID: PMC6182186
- DOI: 10.1093/nar/gky761
The somatic piRNA pathway controls germline transposition over generations
Abstract
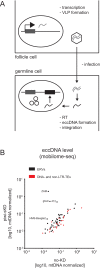
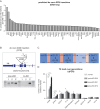
Transposable elements (TEs) are parasitic DNA sequences that threaten genome integrity by replicative transposition in host gonads. The Piwi-interacting RNAs (piRNAs) pathway is assumed to maintain Drosophila genome homeostasis by downregulating transcriptional and post-transcriptional TE expression in the ovary. However, the bursts of transposition that are expected to follow transposome derepression after piRNA pathway impairment have not yet been reported. Here, we show, at a genome-wide level, that piRNA loss in the ovarian somatic cells boosts several families of the endogenous retroviral subclass of TEs, at various steps of their replication cycle, from somatic transcription to germinal genome invasion. For some of these TEs, the derepression caused by the loss of piRNAs is backed up by another small RNA pathway (siRNAs) operating in somatic tissues at the post transcriptional level. Derepressed transposition during 70 successive generations of piRNA loss exponentially increases the genomic copy number by up to 10-fold.
Figures


References
-
- Jurka J., Kapitonov V.V., Kohany O., Jurka M.V.. Repetitive sequences in complex genomes: structure and evolution. Annu. Rev. Genomics Hum. Genet. 2007; 8:241–259. - PubMed
-
- Finnegan D.J. Transposable elements and DNA transposition in eukaryotes. Curr. Opin. Cell Biol. 1990; 2:471–477. - PubMed
-
- Anxolabéhère D., Kidwell M.G., Periquet G.. Molecular characteristics of diverse populations are consistent with the hypothesis of a recent invasion of Drosophila melanogaster by mobile P elements. Mol. Biol. Evol. 1988; 5:252–269. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

