CEACAM1 in Liver Injury, Metabolic and Immune Regulation
- PMID: 30314283
- PMCID: PMC6213298
- DOI: 10.3390/ijms19103110
CEACAM1 in Liver Injury, Metabolic and Immune Regulation
Abstract
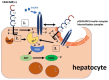
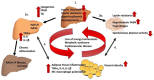
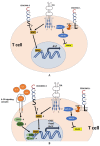
Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) is a transmembrane glycoprotein that is expressed on epithelial, endothelial and immune cells. CEACAM1 is a differentiation antigen involved in the maintenance of epithelial polarity that is induced during hepatocyte differentiation and liver regeneration. CEACAM1 regulates insulin sensitivity by promoting hepatic insulin clearance, and controls liver tolerance and mucosal immunity. Obese insulin-resistant humans with non-alcoholic fatty liver disease manifest loss of hepatic CEACAM1. In mice, deletion or functional inactivation of CEACAM1 impairs insulin clearance and compromises metabolic homeostasis which initiates the development of obesity and hepatic steatosis and fibrosis with other features of non-alcoholic steatohepatitis, and adipogenesis in white adipose depot. This is followed by inflammation and endothelial and cardiovascular dysfunctions. In obstructive and inflammatory liver diseases, soluble CEACAM1 is shed into human bile where it can serve as an indicator of liver disease. On immune cells, CEACAM1 acts as an immune checkpoint regulator, and deletion of Ceacam1 gene in mice causes exacerbation of inflammation and hyperactivation of myeloid cells and lymphocytes. Hence, hepatic CEACAM1 resides at the central hub of immune and metabolic homeostasis in both humans and mice. This review focuses on the regulatory role of CEACAM1 in liver and biliary tract architecture in health and disease, and on its metabolic role and function as an immune checkpoint regulator of hepatic inflammation.
Keywords: CEACAM1; immune checkpoint receptor; insulin clearance; liver disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Hinoda Y., Neumaier M., Hefta S.A., Drzeniek Z., Wagener C., Shively L., Hefta L.J., Shively J.E., Paxton R.J. Molecular cloning of a cDNA coding biliary glycoprotein I: Primary structure of a glycoprotein immunologically crossreactive with carcinoembryonic antigen. Proc. Natl. Acad. Sci. USA. 1988;85:6959–6963. doi: 10.1073/pnas.85.18.6959. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

