ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival
- PMID: 30314966
- PMCID: PMC6213280
- DOI: 10.15252/embj.201899753
ESCRT-mediated lysosome repair precedes lysophagy and promotes cell survival
Abstract
Although lysosomes perform a number of essential cellular functions, damaged lysosomes represent a potential hazard to the cell. Such lysosomes are therefore engulfed by autophagic membranes in the process known as lysophagy, which is initiated by recognition of luminal glycoprotein domains by cytosolic lectins such as Galectin-3. Here, we show that, under various conditions that cause injury to the lysosome membrane, components of the endosomal sorting complex required for transport (ESCRT)-I, ESCRT-II, and ESCRT-III are recruited. This recruitment occurs before that of Galectin-3 and the lysophagy machinery. Subunits of the ESCRT-III complex show a particularly prominent recruitment, which depends on the ESCRT-I component TSG101 and the TSG101- and ESCRT-III-binding protein ALIX Interference with ESCRT recruitment abolishes lysosome repair and causes otherwise reversible lysosome damage to become cell lethal. Vacuoles containing the intracellular pathogen Coxiella burnetii show reversible ESCRT recruitment, and interference with this recruitment reduces intravacuolar bacterial replication. We conclude that the cell is equipped with an endogenous mechanism for lysosome repair which protects against lysosomal damage-induced cell death but which also provides a potential advantage for intracellular pathogens.
Keywords: autophagy; endosome; lysophagy; lysosome; membrane repair.
© 2018 The Authors.
Figures
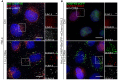
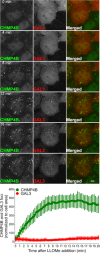
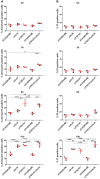
Representative fluorescence micrographs of HeLa cells treated with 250 μM lysosomotropic drug LLOMe or equal volume of DMSO (Ctrl) for 1 h before fixation and immunostained with Hoechst (blue), anti‐CHMP4B (green), anti‐GAL3 (red), and anti‐LAMP1 (white) are presented. Cells treated with LLOMe show increased recruitment of ESCRT‐III protein to lysosomes when compared to Ctrl cells. Number of foci per cell was quantified and is indicated as mean ± SD. Data are quantified from >86 cells per condition from three independent experiments. CHMP4B, GAL3, LAMP1 foci per cell (Ctrl versus LLOMe treatment): P = 0.0398, P = 0.0033, P = 0.2400 (Student's t‐test), respectively.
HeLa cells stably expressing CHMP4B‐eGFP and mCherry‐Galectin‐3 treated as in (A) and stained with CD63 antibody are shown. Number of foci per cell was quantified and is indicated as mean ± SD. Data are quantified from >87 cells per condition from three independent experiments. CHMP4B, GAL3, CD63 foci per cell (Ctrl versus LLOMe treatment): P = 0.0111, P = 0.0050, P = 0.3793 (Student's t‐test), respectively.

Representative fluorescence images of a HeLa‐mCherry‐Galectin‐3 stable cell line treated with 250 μM lysosomotropic drug LLOMe for 1 h, fixed and labeled with CD63 antibody. To determine the statistical significance, the number of CHMP4B, GAL3, and CD63 foci per cell observed after LLOMe treatment was compared to Ctrl using Student's t‐test. P‐values for CHMP4B, GAL3, and CD63 are as follows: P = 0.000001, P = 0.0017, P = 0.4160, respectively.
Experimental setup as in (A) where RPE‐1 cells are labeled with CHMP4B, Galectin‐3, and LAMP1 antibodies. P‐values for CHMP4B, GAL3, and LAMP1 foci per cell (Ctrl versus LLOMe treatment): P = 0.00001, P = 0.00018, P = 0.1368 (Student's t‐test), respectively.
Representative images of fixed H‐460 cells after 1‐h treatment with 250 μM LLOMe. Cells were stained for endogenous CHMP4B, GAL3, and LAMP1. CHMP4B, GAL3, and LAMP1 foci per cell (Ctrl versus LLOMe treatment): P = 0.0192, P = 0.0493, P = 0.6333 (Student's t‐test), respectively.

HeLa cells stably expressing CHMP4B‐eGFP and mCherry‐Galectin‐3 were treated with 250 μM lysosomotropic drug GPN for 1 h, fixed and imaged. Representative images showing recruitment of CHMP4B to damaged endolysosomal membranes are presented. Number of foci per cell (>90 cells per condition from four independent experiments) was quantified and is indicated as mean ± SD. P‐values for comparisons CHMP4B, GAL3, and LAMP1 foci per cell (Ctrl versus GPN treatment) are as follows: P = 0.0002, P = 0.0008, P = 0.1113 (Student's t‐test), respectively.
Representative fluorescence images showing recruitment of CHMP4B to damaged membranes in HeLa cells stably expressing CHMP4B‐eGFP and mCherry‐Galectin‐3 and RPE‐1 cells after 2‐h treatment with 7 μm cationic amphiphilic drug terfenadine. In HeLa cells, there is a slight but not significant increase in CHMP4B‐positive foci. On the other hand, RPE‐1 cells labeled with CHMP4B, GAL3, and LAMP1 antibodies have elevated number of CHMP4 and GAL3 foci per cell. For HeLa‐CHMP4BeGFP‐mCherry‐Galectin‐3: CHMP4B, GAL3 foci per cell (Ctrl versus terfenadine treatment): P = 0.2679, P = 0.2266 (Student's t‐test), respectively. For RPE‐1 cell line: CHMP4B, GAL3 foci per cell (Ctrl versus terfenadine treatment): P = 0.0169, P = 0.1806 (Student's t‐test), respectively. Number of foci per cell was quantified from >75 cells per condition from two independent experiments and is indicated in the figure as mean ± SD.
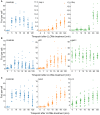
Depletion of HRS does not alter the dynamics of CHMP4B‐eGFP recruitment as compared to siCtrl during treatment with LLOMe.
Downregulation of TSG101, with two independent siRNAs, causes a delay in CHMP4B‐eGFP recruitment as compared to siCtrl, indicating an important role of the ESCRT‐I complex in recruiting the downstream components.
CHMP2A depletion accumulates and stabilizes CHMP4B‐eGFP‐positive foci.
siRNA‐mediated depletion of ALIX shows no significant change on dynamics of CHMP4B recruitment.
Simultaneous depletion of TSG101 and ALIX caused almost no recruitment of CHMP4B upon induction of endolysosomal damage.
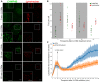
HeLa cells stably expressing CHMP4B‐eGFP were treated with 250 μM LLOMe and 75 nM Lysotracker DND‐99 and fixed at different time points as indicated. After 10 min of LLOMe treatment, CHMP4B is recruited whereas the number of Lysotracker‐positive foci is reduced. After 30 min, lysosomes gain back functionality (judging by the increased number of Lysotracker foci) and appear recovered after 1 h indicating that the ESCRT complex is able to seal the damaged endolysosomal membranes. Representative confocal images for each time point are shown. Scale bars: 5 μm.
Quantification graph (>250 cells per condition from four independent experiments) showing CHMP4B and Lysotracker‐positive foci per cell at different time points. Error bars correspond to 95% confidence intervals.
Quantification graph showing dynamics of Lysotracker recovery in control (siCtrl) and both TSG101‐ and ALIX‐depleted cells. HeLa cells stably expressing CHMP4B‐eGFP were co‐transfected with siRNAs against TSG101 and ALIX. Forty‐eight hours post‐transfection, cells were pre‐treated for 20 min with 75 nM Lysotracker Deep Red, which was used as a read‐out. While the decrease in the number of Lysotracker spots is quickly recovered in siCtrl, simultaneous depletion of ALIX and TSG101 leads to a severe impairment in lysosomal repair. Data from >40 cells per condition from four independent live‐cell imaging experiments are shown. Graph is normalized to the area occupied by the cells. Error bars correspond to 95% confidence intervals.
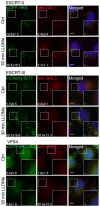
HeLa cells stably expressing CHMP4B‐eGFP and mCherry‐Galectin‐3 were incubated with 250 μM LLOMe, fixed at different time points as indicated and stained for ubiquitin. As presented, CHMP4B is recruited before ubiquitin and Galectin‐3 upon lysosomal membrane damage.
HeLa cells stably expressing CHMP4B‐eGFP were treated as in (A) and stained for p62. CHMP4B is recruited before p62 on the damaged lysosomes. The number of LAMP1‐positive foci appears stable independently of LLOMe treatment.
Experimental setup as in (A and B) where HeLa cells stably expressing CHMP4B‐eGFP and mCherry‐Galectin‐3 were stained for LC3. As shown, CHMP4B is recruited prior to LC3.
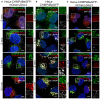
- A
Recruitment of CHMP4B as early as 5 min following LLOMe treatment whereas ubiquitin appears to be recruited later.
- B, C
Recruitment of CHMP4B to damaged endolysosomal membranes precedes p62 and LC3. In addition, p62 is recruited before LC3.
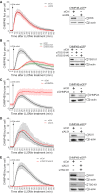
HeLa cells were transfected with different siRNAs as indicated. Forty‐eight hours post‐transfection, cells were treated with 250 μM LLOMe for 3, 6, and 10 h harvested and stained for Annexin V. Cells were immediately processed for flow cytometry. Already after 3 h of treatment with LLOMe, cells co‐transfected with TSG101+ALIX siRNA showed an increase in apoptotic cell death, whereas TSG101 siRNA‐transfected cells showed elevated Annexin V staining after 6 h of treatment.
Experimental set up as in (A) with additional propidium iodide (PI) staining of transfected HeLa cells.
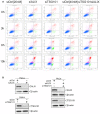
Cells were transfected with control (siCtrl), TSG101, ALIX, or both siRNAs. Forty‐eight hours after transfection, cells were harvested and profiled with flow cytometry. Dot plots and gating strategy of one out of three experiments are shown.
HeLa cells were transfected with control (siCtrl), ALIX, TSG101, or both siRNA. Efficient depletion was confirmed by Western blot analysis. β‐actin was used as loading control.
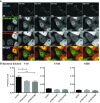
HeLa cells stably expressing CHMP4B‐eGFP were transfected with the mCherry‐Galectin‐3 plasmid. Twenty‐four hours later, cells were infected with WT Coxiella burnetii. Live‐cell imaging started 24 h post‐infection, and several time points are indicated during the next 24 h of observation (time indicated in minutes, see Movie EV9). Arrows indicate Coxiella‐containing vacuole (DRAQ5 labeling) becoming positive for CHMP4B‐eGFP and mCherry‐Galectin‐3. Scale bars: 50 μm.
Bacterial viability and replication assay upon TSG101 KD. HeLa cells were treated with either siCtrl or with two different siTSG101 for 48 h. Then, cells were infected with mCherry‐Coxiella burnetii for 48 h before lysis and serial dilutions were made to infect Vero cells. Seventy‐two hours later, infected cells were fixed, DAPI stained, and processed for quantitative image analysis. Between 30 and 40 fields representing more than 9,000 cells per condition of three independent experiments were analyzed. Error bars represent SD. Statistical significance was determined using one‐way ANOVA test. **P ≤ 0.01 and ***P ≤ 0.001.
References
-
- Babst M, Katzmann DJ, Estepa‐Sabal EJ, Meerloo T, Emr SD (2002a) Escrt‐III: an endosome‐associated heterooligomeric protein complex required for mvb sorting. Dev Cell 3: 271–282 - PubMed
-
- Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD (2002b) Endosome‐associated complex, ESCRT‐II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell 3: 283–289 - PubMed
-
- Bissig C, Gruenberg J (2014) ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol 24: 19–25 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

