Increased sequence hydrophobicity reduces conformational specificity: A mutational case study of the Arc repressor protein
- PMID: 30315592
- PMCID: PMC6322834
- DOI: 10.1002/prot.25613
Increased sequence hydrophobicity reduces conformational specificity: A mutational case study of the Arc repressor protein
Abstract
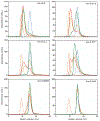
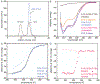
The amino-acid sequences of soluble, globular proteins must have hydrophobic residues to form a stable core, but excess sequence hydrophobicity can lead to loss of native state conformational specificity and aggregation. Previous studies of polar-to-hydrophobic mutations in the β-sheet of the Arc repressor dimer showed that a single substitution at position 11 (N11L) leads to population of an alternate dimeric fold in which the β-sheet is replaced by helix. Two additional hydrophobic mutations at positions 9 and 13 (Q9V and R13V) lead to population of a differently folded octamer along with both dimeric folds. Here we conduct a comprehensive study of the sequence determinants of this progressive loss of fold specificity. We find that the alternate dimer-fold specifically results from the N11L substitution and is not promoted by other hydrophobic substitutions in the β-sheet. We also find that three highly hydrophobic substitutions at positions 9, 11, and 13 are necessary and sufficient for oligomer formation, but the oligomer size depends on the identity of the hydrophobic residue in question. The hydrophobic substitutions increase thermal stability, illustrating how increased hydrophobicity can increase folding stability even as it degrades conformational specificity. The oligomeric variants are predicted to be aggregation-prone but may be hindered from doing so by proline residues that flank the β-sheet region. Loss of conformational specificity due to increased hydrophobicity can manifest itself at any level of structure, depending upon the specific mutations and the context in which they occur.
Keywords: conformational specificity; protein conformation; protein folding; sequence hydrophobicity; sequence-structure relationship; structural degeneracy.
© 2018 Wiley Periodicals, Inc.
Figures
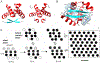
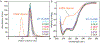
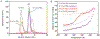

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

