RNA viromes of the oriental hybrid lily cultivar "Sorbonne"
- PMID: 30316297
- PMCID: PMC6186116
- DOI: 10.1186/s12864-018-5138-3
RNA viromes of the oriental hybrid lily cultivar "Sorbonne"
Abstract
Background: The lily is a perennial flowering plant belonging to the genus Lilium in the family Liliaceae. Most cultivated lily plants are propagated by bulbs. Therefore, numerous lily bulbs are frequently infected by diverse viruses causing viral diseases. To date, no study has examined the viromes of plants of one type with identical genetic backgrounds collected from different geographical regions.
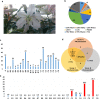
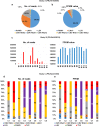
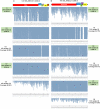
Results: Here, we examined different viromes of the lily cultivar "Sorbonne" using 172 gigabytes of transcriptome data composed of 23 libraries from four different projects for the cultivar "Sorbonne." We identified 396 virus-associated contigs from all but one library. We identified six different viruses, including Plantago asiatica mosaic virus (PlAMV), Cucumber mosaic virus (CMV), Lily symptomless virus (LSV), Tulip virus X (TVX), Lily mottle virus (LMoV), and Tobacco rattle virus (TRV). Of them, PlAMV was the most common virus infecting the lily. Scale and flower samples possessed a high number of virus-associated reads. We assembled 32 nearly complete genomes for the six identified viruses possessing the polyadenylate tails. Genomes of all six viruses were highly conserved in the lily cultivar "Sorbonne" based on mutation analysis. We identified defective RNAs from LSV, TVX, and PlAMV localized in the triple gene block region. Phylogenetic analyses showed that virus genomes are highly correlated with geographical regions and host plants.
Conclusions: We conducted comprehensive virome analyses of a single lily cultivar, "Sorbonne," using transcriptome data. Our results shed light on an array of lily virome-associated topics, including virus identification, the dominant virus, virus accumulation in different plant tissues, virus genome assembly, virus mutation, identification of defective RNAs, and phylogenetic relationships of identified viruses. Taken together, we provide very useful methods and valuable results that can be applied in other virome-associated studies.
Keywords: Genome; Lily; Transcriptome; Virome; Virus; “Sorbonne”.
Conflict of interest statement
Ethics approval and consent to participate
This study did not include the use of any animals, human or otherwise, so did not require ethical approval.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures






References
-
- Lim K-B, Barba-Gonzalez R, Zhou S, Ramanna M, Van Tuyl JM. Interspecific hybridization in lily (Lilium): taxonomic and commercial aspects of using species hybrids in breeding. Floriculture Ornamental Plant biotechnol. 2008;5:138–145.
-
- Simmonds J, Cumming B. Propagation of Lilium hybrids. II. Production of plantlets from bulb-scale callus cultures for increased propagation rates. Sci Hortic. 1976;5(2):161–170. doi: 10.1016/0304-4238(76)90078-9. - DOI
-
- Chastagner Gary A., van Tuyl Jaap M., Verbeek Martin, Miller William B., Westerdahl Becky B. Handbook of Plant Disease Management. Cham: Springer International Publishing; 2017. Diseases of Lily; pp. 1–61.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

