An insight into gut microbiota and its functionalities
- PMID: 30317530
- PMCID: PMC11105460
- DOI: 10.1007/s00018-018-2943-4
An insight into gut microbiota and its functionalities
Abstract
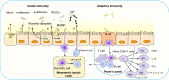
Gut microbiota has evolved along with their hosts and is an integral part of the human body. Microbiota acquired at birth develops in parallel as the host develops and maintains its temporal stability and diversity through adulthood until death. Recent developments in genome sequencing technologies, bioinformatics and culturomics have enabled researchers to explore the microbiota and in particular their functions at more detailed level than before. The accumulated evidences suggest that though a part of the microbiota is conserved, the dynamic members vary along the gastrointestinal tract, from infants to elderly, primitive tribes to modern societies and in different health conditions. Though the gut microbiota is dynamic, it performs some basic functions in the immunological, metabolic, structural and neurological landscapes of the human body. Gut microbiota also exerts significant influence on both physical and mental health of an individual. An in-depth understanding of the functioning of gut microbiota has led to some very exciting developments in therapeutics, such as prebiotics, probiotics, drugs and faecal transplantation leading to improved health.
Keywords: Functions; Gut microbiota; Health; Therapeutics.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

