Nanoparticles Self-Assembly within Lipid Bilayers
- PMID: 30320248
- PMCID: PMC6173477
- DOI: 10.1021/acsomega.8b01445
Nanoparticles Self-Assembly within Lipid Bilayers
Abstract
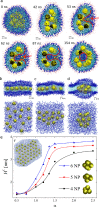
Coarse-grained molecular dynamics simulations are used to model the self-assembly of small hydrophobic nanoparticles (NPs) within the interior of lipid bilayers. The simulation results reveal the conditions under which NPs form clusters and lattices within lipid bilayers of planar and spherical shapes, depending on the NP-lipid coupling strengths. The formation of nanopores within spherical bilayers with self-assembled planar NPs is also described. These observations can provide guidance in the preparation of functional bio-inorganic systems.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Zhang S. Building from the bottom up. Mater. Today 2003, 6, 20–27. 10.1016/s1369-7021(03)00530-3. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

