Conformational Sampling of Macrocyclic Drugs in Different Environments: Can We Find the Relevant Conformations?
- PMID: 30320271
- PMCID: PMC6173504
- DOI: 10.1021/acsomega.8b01379
Conformational Sampling of Macrocyclic Drugs in Different Environments: Can We Find the Relevant Conformations?
Abstract
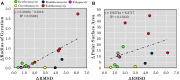
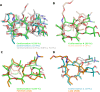
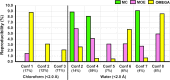
Conformational flexibility is a major determinant of the properties of macrocycles and other drugs in beyond rule of 5 (bRo5) space. Prediction of conformations is essential for design of drugs in this space, and we have evaluated three tools for conformational sampling of a set of 10 bRo5 drugs and clinical candidates in polar and apolar environments. The distance-geometry based OMEGA was found to yield ensembles spanning larger structure and property spaces than the ensembles obtained by MOE-LowModeMD (MOE) and MacroModel (MC). Both MC and OMEGA but not MOE generated different ensembles for polar and apolar environments. All three conformational search methods generated conformers similar to the crystal structure conformers for 9 of the 10 compounds, with OMEGA performing somewhat better than MOE and MC. MOE and OMEGA found all six conformers of roxithromycin that were identified by NMR in aqueous solutions, whereas only OMEGA sampled the three conformers observed in chloroform. We suggest that characterization of conformers using molecular descriptors, e.g., the radius of gyration and polar surface area, is preferred to energy- or root-mean-square deviation-based methods for selection of biologically relevant conformers in drug discovery in bRo5 space.
Conflict of interest statement
The authors declare no competing financial interest.
Figures