Clinical Use of Precision Oncology Decision Support
- PMID: 30320296
- PMCID: PMC6179148
- DOI: 10.1200/PO.17.00036
Clinical Use of Precision Oncology Decision Support
Abstract
Purpose: Precision oncology is hindered by the lack of decision support for determining the functional and therapeutic significance of genomic alterations in tumors and relevant clinically available options. To bridge this knowledge gap, we established a Precision Oncology Decision Support (PODS) team that provides annotations at the alteration-level and subsequently determined if clinical decision-making was influenced.
Methods: Genomic alterations were annotated to determine actionability based on a variant's known or potential functional and/or therapeutic significance. The medical records of a subset of patients annotated in 2015 were manually reviewed to assess trial enrollment. A web-based survey was implemented to capture the reasons why genotype-matched therapies were not pursued.
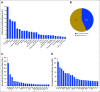
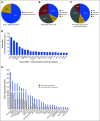
Results: PODS processed 1,669 requests for annotation of 4,084 alterations (2,254 unique) across 49 tumor types for 1,197 patients. 2,444 annotations for 669 patients included an actionable variant call: 32.5% actionable, 9.4% potentially, 29.7% unknown, 28.4% non-actionable. 66% of patients had at least one actionable/potentially actionable alteration. 20.6% (110/535) patients annotated enrolled on a genotype-matched trial. Trial enrolment was significantly higher for patients with actionable/potentially actionable alterations (92/333, 27.6%) than those with unknown (16/136, 11.8%) and non-actionable (2/66, 3%) alterations (p=0.00004). Actionable alterations in PTEN, PIK3CA, and ERBB2 most frequently led to enrollment on genotype-matched trials. Clinicians cited a variety of reasons why patients with actionable alterations did not enroll on trials.
Conclusion: Over half of alterations annotated were of unknown significance or non-actionable. Physicians were more likely to enroll a patient on a genotype-matched trial when an annotation supported actionability. Future studies are needed to demonstrate the impact of decision support on trial enrollment and oncologic outcomes.
Conflict of interest statement
Declaration of Interests: Amber Johnson, Yekaterina Khotskaya, Lauren Brusco, Jia Zeng, Vijaykumar Holla, Ann Bailey, Beate Lizenburger, Nora Sanchez, Md Abu Shufean, Sarina Piha-Paul, Vivek Subbiah, David Hong, Mark Routbort, Russell Broaddus, and Kenna Shaw declare no conflict of interest. Dr. Funda Meric-Bernstam is conducting research sponsored by Aileron, AstraZeneca, Bayer, Calithera, CytoMx, Effector Pharmaceuticals, Debiopharma, Genentech, Novartis, PUMA, Taiho, and Zymeworks. She serves as a consultant for Dialecta and is a member on the Advisory Boards for Clearlight Diagnostics, Darwin Health, GRAIL, Inflection Biosciences, and Pieris. Dr. Gordon Mills serves as a consultant for Adventist Health, Allostery, AstraZeneca, Catena Pharmaceuticals, Critical Outcome Technologies, ImmunoMet, Lilly, Medimmune, Nuevolution, Novartis, Precision Medicine, Provista Diagnostics, Roche, Signalchem Lifesciences, Symphogen, Takeda/Millenium Parmaceuticals, Tau Therapeutics, and Tarveda. Dr. Mills holds stock options in Catena Pharmaceuticals, ImmunoMet, and Spindle Top Ventures. Dr. Mills is conducting research sponsored by Adelson Medical Research Foundation, AstraZeneca, Breast Cancer Research Foundation, Critical Outcomes Technology, Illumina, Ionis, Karus, Komen Research Foundation, Nanostring, and Takeda/Millenium Pharmaceuticals. Dr. John Mendelsohn is a compensated board member at Merrimack Pharmaceuticals, where he also owns stock. He also receives royalty payments from the University of California San Diego.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

