New approach for understanding genome variations in KEGG
- PMID: 30321428
- PMCID: PMC6324070
- DOI: 10.1093/nar/gky962
New approach for understanding genome variations in KEGG
Abstract
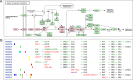
KEGG (Kyoto Encyclopedia of Genes and Genomes; https://www.kegg.jp/ or https://www.genome.jp/kegg/) is a reference knowledge base for biological interpretation of genome sequences and other high-throughput data. It is an integrated database consisting of three generic categories of systems information, genomic information and chemical information, and an additional human-specific category of health information. KEGG pathway maps, BRITE hierarchies and KEGG modules have been developed as generic molecular networks with KEGG Orthology nodes of functional orthologs so that KEGG pathway mapping and other procedures can be applied to any cellular organism. Unfortunately, however, this generic approach was inadequate for knowledge representation in the health information category, where variations of human genomes, especially disease-related variations, had to be considered. Thus, we have introduced a new approach where human gene variants are explicitly incorporated into what we call 'network variants' in the recently released KEGG NETWORK database. This allows accumulation of knowledge about disease-related perturbed molecular networks caused not only by gene variants, but also by viruses and other pathogens, environmental factors and drugs. We expect that KEGG NETWORK will become another reference knowledge base for the basic understanding of disease mechanisms and practical use in clinical sequencing and drug development.
Figures





Similar articles
-
KEGG: new perspectives on genomes, pathways, diseases and drugs.Nucleic Acids Res. 2017 Jan 4;45(D1):D353-D361. doi: 10.1093/nar/gkw1092. Epub 2016 Nov 28. Nucleic Acids Res. 2017. PMID: 27899662 Free PMC article.
-
KEGG Bioinformatics Resource for Plant Genomics and Metabolomics.Methods Mol Biol. 2016;1374:55-70. doi: 10.1007/978-1-4939-3167-5_3. Methods Mol Biol. 2016. PMID: 26519400
-
KEGG: integrating viruses and cellular organisms.Nucleic Acids Res. 2021 Jan 8;49(D1):D545-D551. doi: 10.1093/nar/gkaa970. Nucleic Acids Res. 2021. PMID: 33125081 Free PMC article.
-
The KEGG database.Novartis Found Symp. 2002;247:91-101; discussion 101-3, 119-28, 244-52. Novartis Found Symp. 2002. PMID: 12539951 Review.
-
Knowledge-Based Analysis of Protein Interaction Networks in Neurodegenerative Diseases.In: Alzate O, editor. Neuroproteomics. Boca Raton (FL): CRC Press/Taylor & Francis; 2010. Chapter 9. In: Alzate O, editor. Neuroproteomics. Boca Raton (FL): CRC Press/Taylor & Francis; 2010. Chapter 9. PMID: 21882442 Free Books & Documents. Review.
Cited by
-
Single-cell analysis of matrisome-related genes in breast invasive carcinoma: new avenues for molecular subtyping and risk estimation.Front Immunol. 2024 Oct 18;15:1466762. doi: 10.3389/fimmu.2024.1466762. eCollection 2024. Front Immunol. 2024. PMID: 39493752 Free PMC article.
-
Well-Differentiated Papillary Mesothelioma of the Peritoneum Is Genetically Distinct from Malignant Mesothelioma.Cancers (Basel). 2020 Jun 13;12(6):1568. doi: 10.3390/cancers12061568. Cancers (Basel). 2020. PMID: 32545767 Free PMC article.
-
Identification of Biomarkers to Construct a Competing Endogenous RNA Network and Establishment of a Genomic-Clinicopathologic Nomogram to Predict Survival for Children with Rhabdoid Tumors of the Kidney.Biomed Res Int. 2020 Aug 26;2020:5843874. doi: 10.1155/2020/5843874. eCollection 2020. Biomed Res Int. 2020. PMID: 32908900 Free PMC article.
-
Drosophila models of pathogenic copy-number variant genes show global and non-neuronal defects during development.PLoS Genet. 2020 Jun 24;16(6):e1008792. doi: 10.1371/journal.pgen.1008792. eCollection 2020 Jun. PLoS Genet. 2020. PMID: 32579612 Free PMC article.
-
Identification and verification of hub genes associated with the progression of non-small cell lung cancer by integrated analysis.Front Pharmacol. 2022 Sep 13;13:997842. doi: 10.3389/fphar.2022.997842. eCollection 2022. Front Pharmacol. 2022. PMID: 36176446 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

