Amide Bond Activation of Biological Molecules
- PMID: 30322008
- PMCID: PMC6222841
- DOI: 10.3390/molecules23102615
Amide Bond Activation of Biological Molecules
Abstract
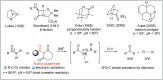
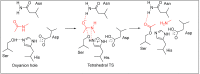
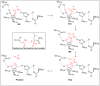
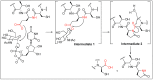
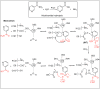
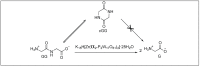
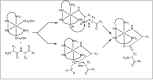
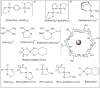
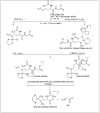
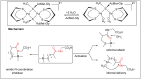
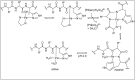
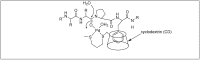
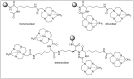
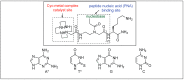
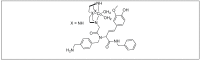
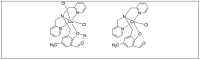
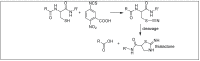
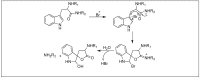
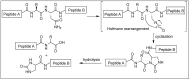
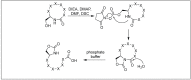
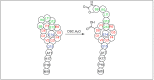
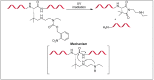
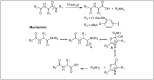
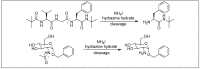
Amide bonds are the most prevalent structures found in organic molecules and various biomolecules such as peptides, proteins, DNA, and RNA. The unique feature of amide bonds is their ability to form resonating structures, thus, they are highly stable and adopt particular three-dimensional structures, which, in turn, are responsible for their functions. The main focus of this review article is to report the methodologies for the activation of the unactivated amide bonds present in biomolecules, which includes the enzymatic approach, metal complexes, and non-metal based methods. This article also discusses some of the applications of amide bond activation approaches in the sequencing of proteins and the synthesis of peptide acids, esters, amides, and thioesters.
Keywords: amide bond resonance; catalysts; enzymes; metal complexes; peptide bond cleavage; twisted amides.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






























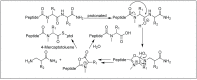

References
-
- Greenberg A., Breneman C.M., Liebman J.F. The Amide Linkage: Structural Significance, Shemistry, Biochemistry and Material Science. Wiley; New York, NY, USA: 2000.
-
- Brunton L., Chabner B., Knollman B. Goodman and Gilman’s the Pharmacological Basis of Therapeutics. MacGraw-Hill; New York, NY, USA: 2010.
-
- Hughes A.B. Amino Acids, Peptides and Proteins in Organic Chemistry. Wiley-VCH; Weinheim, Germany: 2009.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

