Role of Extracellular Loops and Membrane Lipids for Ligand Recognition in the Neuronal Adenosine Receptor Type 2A: An Enhanced Sampling Simulation Study
- PMID: 30322034
- PMCID: PMC6222423
- DOI: 10.3390/molecules23102616
Role of Extracellular Loops and Membrane Lipids for Ligand Recognition in the Neuronal Adenosine Receptor Type 2A: An Enhanced Sampling Simulation Study
Abstract
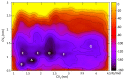
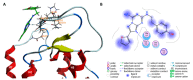
Human G-protein coupled receptors (GPCRs) are important targets for pharmaceutical intervention against neurological diseases. Here, we use molecular simulation to investigate the key step in ligand recognition governed by the extracellular domains in the neuronal adenosine receptor type 2A (hA2AR), a target for neuroprotective compounds. The ligand is the high-affinity antagonist (4-(2-(7-amino-2-(furan-2-yl)-[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-ylamino)ethyl)phenol), embedded in a neuronal membrane mimic environment. Free energy calculations, based on well-tempered metadynamics, reproduce the experimentally measured binding affinity. The results are consistent with the available mutagenesis studies. The calculations identify a vestibular binding site, where lipids molecules can actively participate to stabilize ligand binding. Bioinformatic analyses suggest that such vestibular binding site and, in particular, the second extracellular loop, might drive the ligand toward the orthosteric binding pocket, possibly by allosteric modulation. Taken together, these findings point to a fundamental role of the interaction between extracellular loops and membrane lipids for ligands' molecular recognition and ligand design in hA2AR.
Keywords: adenosine receptor; allosterism; extracellular loops; metadynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Fink J.S., Weaver D.R., Rivkees S.A., Peterfreund R.A., Pollack A.E., Adler E.M., Reppert S.M. Molecular cloning of the rat A2 adenosine receptor: Selective co-expression with D2 dopamine receptors in rat striatum. Brain Res. Mol. Brain Res. 1992;14:186–195. doi: 10.1016/0169-328X(92)90173-9. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

