Mitochondrial Arabidopsis thaliana TRXo Isoforms Bind an Iron⁻Sulfur Cluster and Reduce NFU Proteins In Vitro
- PMID: 30322144
- PMCID: PMC6210436
- DOI: 10.3390/antiox7100142
Mitochondrial Arabidopsis thaliana TRXo Isoforms Bind an Iron⁻Sulfur Cluster and Reduce NFU Proteins In Vitro
Abstract
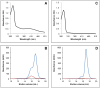
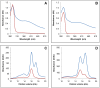
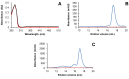
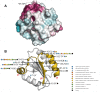
In plants, the mitochondrial thioredoxin (TRX) system generally comprises only one or two isoforms belonging to the TRX h or o classes, being less well developed compared to the numerous isoforms found in chloroplasts. Unlike most other plant species, Arabidopsis thaliana possesses two TRXo isoforms whose physiological functions remain unclear. Here, we performed a structure⁻function analysis to unravel the respective properties of the duplicated TRXo1 and TRXo2 isoforms. Surprisingly, when expressed in Escherichia coli, both recombinant proteins existed in an apo-monomeric form and in a homodimeric iron⁻sulfur (Fe-S) cluster-bridged form. In TRXo2, the [4Fe-4S] cluster is likely ligated in by the usual catalytic cysteines present in the conserved Trp-Cys-Gly-Pro-Cys signature. Solving the three-dimensional structure of both TRXo apo-forms pointed to marked differences in the surface charge distribution, notably in some area usually participating to protein⁻protein interactions with partners. However, we could not detect a difference in their capacity to reduce nitrogen-fixation-subunit-U (NFU)-like proteins, NFU4 or NFU5, two proteins participating in the maturation of certain mitochondrial Fe-S proteins and previously isolated as putative TRXo1 partners. Altogether, these results suggest that a novel regulation mechanism may prevail for mitochondrial TRXs o, possibly existing as a redox-inactive Fe-S cluster-bound form that could be rapidly converted in a redox-active form upon cluster degradation in specific physiological conditions.
Keywords: iron–sulfur cluster; mitochondria; redox regulation; thioredoxin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Navrot N., Rouhier N., Gelhaye E., Jacquot J.P. Reactive oxygen species generation and antioxidant systems in plant mitochondria. Physiol. Plant. 2007;129:185–195. doi: 10.1111/j.1399-3054.2006.00777.x. - DOI
-
- Navrot N., Collin V., Gualberto J., Gelhaye E., Hirasawa M., Rey P., Knaff D.B., Issakidis E., Jacquot J.P., Rouhier N. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 2006;142:1364–1379. doi: 10.1104/pp.106.089458. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

