A comparative study of endoderm differentiation in humans and chimpanzees
- PMID: 30322406
- PMCID: PMC6191992
- DOI: 10.1186/s13059-018-1490-5
A comparative study of endoderm differentiation in humans and chimpanzees
Abstract
Background: There is substantial interest in the evolutionary forces that shaped the regulatory framework in early human development. Progress in this area has been slow because it is difficult to obtain relevant biological samples. Induced pluripotent stem cells (iPSCs) may provide the ability to establish in vitro models of early human and non-human primate developmental stages.
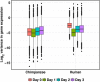
Results: Using matched iPSC panels from humans and chimpanzees, we comparatively characterize gene regulatory changes through a four-day time course differentiation of iPSCs into primary streak, endoderm progenitors, and definitive endoderm. As might be expected, we find that differentiation stage is the major driver of variation in gene expression levels, followed by species. We identify thousands of differentially expressed genes between humans and chimpanzees in each differentiation stage. Yet, when we consider gene-specific dynamic regulatory trajectories throughout the time course, we find that at least 75% of genes, including nearly all known endoderm developmental markers, have similar trajectories in the two species. Interestingly, we observe a marked reduction of both intra- and inter-species variation in gene expression levels in primitive streak samples compared to the iPSCs, with a recovery of regulatory variation in endoderm progenitors.
Conclusions: The reduction of variation in gene expression levels at a specific developmental stage, paired with overall high degree of conservation of temporal gene regulation, is consistent with the dynamics of a conserved developmental process.
Keywords: Comparative genomics; Functional genomics; Gene expression.
Conflict of interest statement
Ethics approval and consent to participate
Human fibroblasts samples for generation of iPSC lines were collected under University of Chicago IRB protocol 11–0524. Informed consent to participate in the study was obtained from all human participants. The original chimpanzee fibroblast samples for generation of iPSC lines were obtained from Yerkes Primate Research Center of Emory University under protocol 006–12, in full accordance with IACUC protocols [12]. All experimental methods comply with the Helsinki Declaration.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures




References
-
- Nowick Katja, Gernat Tim, Almaas Eivind, Stubbs Lisa. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proceedings of the National Academy of Sciences. 2009;106(52):22358–22363. doi: 10.1073/pnas.0911376106. - DOI - PMC - PubMed
-
- Guohua Xu Augix, He Liu, Li Zhongshan, Xu Ying, Li Mingfeng, Fu Xing, Yan Zheng, Yuan Yuan, Menzel Corinna, Li Na, Somel Mehmet, Hu Hao, Chen Wei, Pääbo Svante, Khaitovich Philipp. Intergenic and Repeat Transcription in Human, Chimpanzee and Macaque Brains Measured by RNA-Seq. PLoS Computational Biology. 2010;6(7):e1000843. doi: 10.1371/journal.pcbi.1000843. - DOI - PMC - PubMed
-
- Somel M., Franz H., Yan Z., Lorenc A., Guo S., Giger T., Kelso J., Nickel B., Dannemann M., Bahn S., Webster M. J., Weickert C. S., Lachmann M., Paabo S., Khaitovich P. Transcriptional neoteny in the human brain. Proceedings of the National Academy of Sciences. 2009;106(14):5743–5748. doi: 10.1073/pnas.0900544106. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

