Integrated culturing, modeling and transcriptomics uncovers complex interactions and emergent behavior in a three-species synthetic gut community
- PMID: 30322445
- PMCID: PMC6237439
- DOI: 10.7554/eLife.37090
Integrated culturing, modeling and transcriptomics uncovers complex interactions and emergent behavior in a three-species synthetic gut community
Erratum in
-
Correction: Integrated culturing, modeling and transcriptomics uncovers complex interactions and emergent behavior in a three-species synthetic gut community.Elife. 2019 Nov 4;8:e53217. doi: 10.7554/eLife.53217. Elife. 2019. PMID: 31682225 Free PMC article.
Abstract
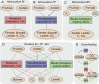
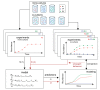
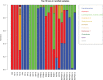
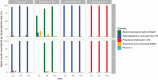
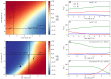
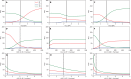
The composition of the human gut microbiome is well resolved, but predictive understanding of its dynamics is still lacking. Here, we followed a bottom-up strategy to explore human gut community dynamics: we established a synthetic community composed of three representative human gut isolates (Roseburia intestinalis L1-82, Faecalibacterium prausnitzii A2-165 and Blautia hydrogenotrophica S5a33) and explored their interactions under well-controlled conditions in vitro. Systematic mono- and pair-wise fermentation experiments confirmed competition for fructose and cross-feeding of formate. We quantified with a mechanistic model how well tri-culture dynamics was predicted from mono-culture data. With the model as reference, we demonstrated that strains grown in co-culture behaved differently than those in mono-culture and confirmed their altered behavior at the transcriptional level. In addition, we showed with replicate tri-cultures and simulations that dominance in tri-culture sensitively depends on the initial conditions. Our work has important implications for gut microbial community modeling as well as for ecological interaction detection from batch cultures.
Keywords: Blautia hydrogenotrophica; Faecalibacterium prausnitzii; Roseburia intestinalis; computational biology; infectious disease; microbiology; systems biology.
© 2018, D'hoe et al.
Conflict of interest statement
KD, SV, KF, FM, GF, DG, VL, LG, JD, LD, JR No competing interests declared
Figures









References
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. GitHub. 2010 https://github.com/s-andrews/FastQC
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

