Regulation of Root Angle and Gravitropism
- PMID: 30322904
- PMCID: PMC6288823
- DOI: 10.1534/g3.118.200540
Regulation of Root Angle and Gravitropism
Abstract
Regulation of plant root angle is critical for obtaining nutrients and water and is an important trait for plant breeding. A plant's final, long-term root angle is the net result of a complex series of decisions made by a root tip in response to changes in nutrient availability, impediments, the gravity vector and other stimuli. When a root tip is displaced from the gravity vector, the short-term process of gravitropism results in rapid reorientation of the root toward the vertical. Here, we explore both short- and long-term regulation of root growth angle, using natural variation in tomato to identify shared and separate genetic features of the two responses. Mapping of expression quantitative trait loci mapping and leveraging natural variation between and within species including Arabidopsis suggest a role for PURPLE ACID PHOSPHATASE 27 and CELL DIVISION CYCLE 73 in determining root angle.
Keywords: gravitropism; root; tomato.
Copyright © 2018 Toal et al.
Figures
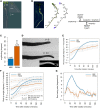
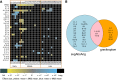



References
-
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., et al. , 2003. Genome-Wide Insertional Mutagenesis of Arabidopsis thaliana. Science 301: 653–657. - PubMed
-
- Andrews S., 2011. FASTQC, pp. A quality control tool for high throughput sequence data, Babraham Institute.
