Single-molecule DNA-mapping and whole-genome sequencing of individual cells
- PMID: 30322920
- PMCID: PMC6217438
- DOI: 10.1073/pnas.1804194115
Single-molecule DNA-mapping and whole-genome sequencing of individual cells
Abstract
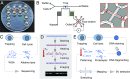
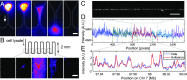
To elucidate cellular diversity and clonal evolution in tissues and tumors, one must resolve genomic heterogeneity in single cells. To this end, we have developed low-cost, mass-producible micro-/nanofluidic chips for DNA extraction from individual cells. These chips have modules that collect genomic DNA for sequencing or map genomic structure directly, on-chip, with denaturation-renaturation (D-R) optical mapping [Marie R, et al. (2013) Proc Natl Acad Sci USA 110:4893-4898]. Processing of single cells from the LS174T colorectal cancer cell line showed that D-R mapping of single molecules can reveal structural variation (SV) in the genome of single cells. In one experiment, we processed 17 fragments covering 19.8 Mb of the cell's genome. One megabase-large fragment aligned well to chromosome 19 with half its length, while the other half showed variable alignment. Paired-end single-cell sequencing supported this finding, revealing a region of complexity and a 50-kb deletion. Sequencing struggled, however, to detect a 20-kb gap that D-R mapping showed clearly in a megabase fragment that otherwise mapped well to the reference at the pericentromeric region of chromosome 4. Pericentromeric regions are complex and show substantial sequence homology between different chromosomes, making mapping of sequence reads ambiguous. Thus, D-R mapping directly, from a single molecule, revealed characteristics of the single-cell genome that were challenging for short-read sequencing.
Keywords: DNA; nanofluidics; optical mapping; sequencing; single cell.
Conflict of interest statement
Conflict of interest statement: R.M., J.N.P., A.K., and K.U.M. filed patents. K.U.M. declares that XGenomes is developing nucleic acid sequencing technologies.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

