mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction
- PMID: 30322972
- PMCID: PMC6279387
- DOI: 10.1083/jcb.201806183
mRNP architecture in translating and stress conditions reveals an ordered pathway of mRNP compaction
Abstract
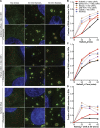
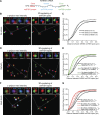
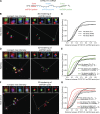
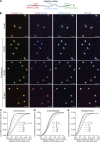
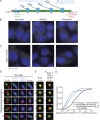
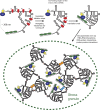
Stress granules (SGs) are transient membraneless organelles of nontranslating mRNA-protein complexes (mRNPs) that form during stress. In this study, we used multiple single-molecule FISH probes for particular mRNAs to examine their SG recruitment and spatial organization. Ribosome runoff is required for SG entry, as long open reading frame (ORF) mRNAs are delayed in SG accumulation, indicating that the SG transcriptome changes over time. Moreover, mRNAs are ∼20× compacted from an expected linear length when translating and compact ∼2-fold further in a stepwise manner beginning at the 5' end during ribosome runoff. Surprisingly, the 5' and 3' ends of the examined mRNAs were separated when translating, but in nontranslating conditions the ends of long ORF mRNAs become close, suggesting that the closed-loop model of mRNPs preferentially forms on nontranslating mRNAs. Compaction of ribosome-free mRNAs is ATP independent, consistent with compaction occurring through RNA structure formation. These results suggest that translation inhibition triggers an mRNP reorganization that brings ends closer, which has implications for the regulation of mRNA stability and translation by 3' UTR elements and the poly(A) tail.
© 2018 Khong and Parker.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

