Processive incorporation of multiple selenocysteine residues is driven by a novel feature of the selenocysteine insertion sequence
- PMID: 30323062
- PMCID: PMC6302164
- DOI: 10.1074/jbc.RA118.005211
Processive incorporation of multiple selenocysteine residues is driven by a novel feature of the selenocysteine insertion sequence
Abstract
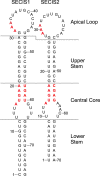
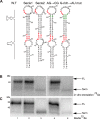
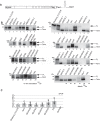
RNA stem loop structures have been frequently shown to regulate essential cellular processes. The selenocysteine insertion sequence (SECIS) element, found in the 3' UTRs of all selenoprotein mRNAs, is an example of such a structure, as it is required for the incorporation of the 21st amino acid, selenocysteine (Sec). Selenoprotein synthesis poses a mechanistic challenge because Sec is incorporated during translation in response to a stop codon (UGA). Although it is known that a SECIS-binding protein (SBP2) is required for Sec insertion, the mechanism of action remains elusive. Additional complexity is present in the synthesis of selenoprotein P (SELENOP), which is the only selenoprotein that contains multiple UGA codons and possesses two SECIS elements in its 3' UTR. Thus, full-length SELENOP synthesis requires processive Sec incorporation. Using zebrafish Selenop, in vitro translation assays, and 75Se labeling in HEK293 cells, we found here that processive Sec incorporation is an intrinsic property of the SECIS elements. Specifically, we identified critical features of SECIS elements that are required for processive Sec incorporation. A screen of the human SECIS elements revealed that most of these elements support processive Sec incorporation in vitro; however, we also found that the processivity of Sec incorporation into Selenop in cells is tightly regulated. We propose a model for processive Sec incorporation that involves differential recruitment of SECIS-binding proteins.
Keywords: 21st amino acid; RNA structure; mRNA; ribonucleoprotein (RNP); selenium; selenocysteine; selenoprotein; stem loop structure.
© 2018 Shetty et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Figures



References
-
- Lee B. J., Worland P. J., Davis J. N., Stadtman T. C., and Hatfield D. L. (1989) Identification of a selenocysteyl-tRNA(Ser) in mammalian cells that recognizes the nonsense codon, UGA. J. Biol. Chem. 264, 9724–9727. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

