Tridimensional infiltration of DNA viruses into the host genome shows preferential contact with active chromatin
- PMID: 30323189
- PMCID: PMC6189100
- DOI: 10.1038/s41467-018-06739-4
Tridimensional infiltration of DNA viruses into the host genome shows preferential contact with active chromatin
Abstract
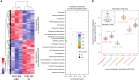
Whether non-integrated viral DNAs distribute randomly or target specific positions within the higher-order architecture of mammalian genomes remains largely unknown. Here we use Hi-C and viral DNA capture (CHi-C) in primary human hepatocytes infected by either hepatitis B virus (HBV) or adenovirus type 5 (Ad5) virus to show that they adopt different strategies in their respective positioning at active chromatin. HBV contacts preferentially CpG islands (CGIs) enriched in Cfp1 a factor required for its transcription. These CGIs are often associated with highly expressed genes (HEG) and genes deregulated during infection. Ad5 DNA interacts preferentially with transcription start sites (TSSs) and enhancers of HEG, as well as genes upregulated during infection. These results show that DNA viruses use different strategies to infiltrate genomic 3D networks and target specific regions. This targeting may facilitate the recruitment of transcription factors necessary for their own replication and contribute to the deregulation of cellular gene expression.
Conflict of interest statement
The authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

