Complex virome in feces from Amerindian children in isolated Amazonian villages
- PMID: 30323210
- PMCID: PMC6189175
- DOI: 10.1038/s41467-018-06502-9
Complex virome in feces from Amerindian children in isolated Amazonian villages
Erratum in
-
Publisher Correction: Complex virome in feces from Amerindian children in isolated Amazonian villages.Nat Commun. 2018 Nov 14;9(1):4852. doi: 10.1038/s41467-018-07337-0. Nat Commun. 2018. PMID: 30429475 Free PMC article.
Abstract
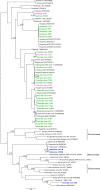
The number of viruses circulating in small isolated human populations may be reduced by viral extinctions and rare introductions. Here we used viral metagenomics to characterize the eukaryotic virome in feces from healthy children from a large urban center and from three Amerindian villages with minimal outside contact. Numerous human enteric viruses, mainly from the Picornaviridae and Caliciviridae families, were sequenced from each of the sites. Multiple children from the same villages shed closely related viruses reflecting frequent transmission clusters. Feces of isolated villagers also contained multiple viral genomes of unknown cellular origin from the Picornavirales order and CRESS-DNA group and higher levels of nematode and protozoan DNA. Despite cultural and geographic isolation, the diversity of enteric human viruses was therefore not reduced in these Amazonian villages. Frequent viral introductions and/or increased susceptibility to enteric infections may account for the complex fecal virome of Amerindian children in isolated villages.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

