A metabolite-derived protein modification integrates glycolysis with KEAP1-NRF2 signalling
- PMID: 30323285
- PMCID: PMC6444936
- DOI: 10.1038/s41586-018-0622-0
A metabolite-derived protein modification integrates glycolysis with KEAP1-NRF2 signalling
Abstract
Mechanisms that integrate the metabolic state of a cell with regulatory pathways are necessary to maintain cellular homeostasis. Endogenous, intrinsically reactive metabolites can form functional, covalent modifications on proteins without the aid of enzymes1,2, and regulate cellular functions such as metabolism3-5 and transcription6. An important 'sensor' protein that captures specific metabolic information and transforms it into an appropriate response is KEAP1, which contains reactive cysteine residues that collectively act as an electrophile sensor tuned to respond to reactive species resulting from endogenous and xenobiotic molecules. Covalent modification of KEAP1 results in reduced ubiquitination and the accumulation of NRF27,8, which then initiates the transcription of cytoprotective genes at antioxidant-response element loci. Here we identify a small-molecule inhibitor of the glycolytic enzyme PGK1, and reveal a direct link between glycolysis and NRF2 signalling. Inhibition of PGK1 results in accumulation of the reactive metabolite methylglyoxal, which selectively modifies KEAP1 to form a methylimidazole crosslink between proximal cysteine and arginine residues (MICA). This posttranslational modification results in the dimerization of KEAP1, the accumulation of NRF2 and activation of the NRF2 transcriptional program. These results demonstrate the existence of direct inter-pathway communication between glycolysis and the KEAP1-NRF2 transcriptional axis, provide insight into the metabolic regulation of the cellular stress response, and suggest a therapeutic strategy for controlling the cytoprotective antioxidant response in several human diseases.
Figures





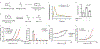


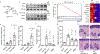
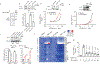


Comment in
-
Playing KEAP1-away.Nat Chem Biol. 2019 Jan;15(1):2. doi: 10.1038/s41589-018-0197-y. Nat Chem Biol. 2019. PMID: 30531902 No abstract available.
References
-
- Weinert BT, et al. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Molecular cell 51, 265–272 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

