Transcription factor dimerization activates the p300 acetyltransferase
- PMID: 30323286
- PMCID: PMC6914384
- DOI: 10.1038/s41586-018-0621-1
Transcription factor dimerization activates the p300 acetyltransferase
Abstract
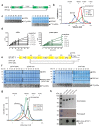
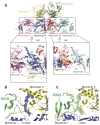
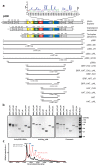
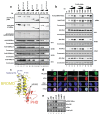
The transcriptional co-activator p300 is a histone acetyltransferase (HAT) that is typically recruited to transcriptional enhancers and regulates gene expression by acetylating chromatin. Here we show that the activation of p300 directly depends on the activation and oligomerization status of transcription factor ligands. Using two model transcription factors, IRF3 and STAT1, we demonstrate that transcription factor dimerization enables the trans-autoacetylation of p300 in a highly conserved and intrinsically disordered autoinhibitory lysine-rich loop, resulting in p300 activation. We describe a crystal structure of p300 in which the autoinhibitory loop invades the active site of a neighbouring HAT domain, revealing a snapshot of a trans-autoacetylation reaction intermediate. Substrate access to the active site involves the rearrangement of an autoinhibitory RING domain. Our data explain how cellular signalling and the activation and dimerization of transcription factors control the activation of p300, and therefore explain why gene transcription is associated with chromatin acetylation.
Conflict of interest statement
The authors declare no competing financial interests.
Figures






References
-
- Parekh BS, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-beta promoter. Mol Cell. 1999;3:125–129. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

