Molecular systematic study of Cardamine glechomifolia Levl. (Brassicaceae) using internal transcribed spacer sequence of nuclear ribosomal DNA (ITS) and chloroplast trn L and trn L-F sequences
- PMID: 30323708
- PMCID: PMC6181152
- DOI: 10.1016/j.sjbs.2010.06.001
Molecular systematic study of Cardamine glechomifolia Levl. (Brassicaceae) using internal transcribed spacer sequence of nuclear ribosomal DNA (ITS) and chloroplast trn L and trn L-F sequences
Abstract
The internal transcribed spacer (ITS) region of nuclear ribosomal DNA, trnL and trnL-F genes of Cardamine glechomifolia Levl. (family Brassicaceae) were sequenced and analyzed with the sequence of related Cardamine species retrieved from NCBI GenBank to detect pattern of evolutionary differentiation. All trees resulting from combined sequence analyses data of ITS, trnL and trnL-F gene resolve that C. glechomifolia - an endemic species to South Korea clade with Cardamine microzyga (100% bootstrap support). The evolutionary history was inferred using the Maximum Parsimony method. The consistency index is (0.588235), the retention index is (0.687500), and the composite index is 0.519622 (0.404412) for all sites and parsimony-informative sites (in parentheses). The result of the analysis using Maximum Parsimony was found congruence with Maximum Likelihood method and in Baseyan analysis.
Keywords: Cardamine glechomifolia; ITS; Molecular systematic; trnL; trnL-F.
Figures


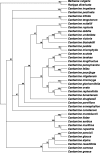


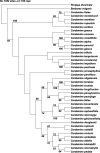
References
-
- Al-Shehbaz I.A. The genera of Arabideae (Cruciferae, Brassicaceae) in the Southeastern United States. J. Arn. Arbor. 1988;69:85–166.
-
- Baldwin B.G. Phylogenetic utility of the internal transcribed spacers of nuclear ribosomal DNA in plants: an example from the Compositae. Mol. Phylo. Evol. 1992;1:3–16. - PubMed
-
- Baldwin B.G., Markos S. Phylogenetic utility of the external transcribed spacer (ETS) of 18S–26S rDNA: congruence of ETS and ITS trees of Calycadenia (Compositae) Mol. Phylo. Evol. 1998;10:449–463. - PubMed
-
- Eck R.V., Dayhoff M.O. National Biomedical Research Foundation; Silver Springs, Maryland: 1966. Atlas of Protein Sequence and Structure.
-
- Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27(2):221–224. - PubMed
LinkOut - more resources
Full Text Sources

